Claire Koenig
@clairekoenig6
PhD student in the @JesperOlsenLab, @UCPH_CPR 👩🏻🔬📓| MSCA PhD fellow @PUSHH_ETN 🦴🔍
ID: 1580185233067999233
12-10-2022 13:15:02
36 Tweet
183 Takipçi
219 Takip Edilen

One of the most recent technological advances in paleontology enables the characterization of ancient proteins, aka #palaeoproteomics. PCI Paleontology reviewed and recommends: Patramanis et al. developed an open-source pipeline, PaleoProPhyler. Check it out! paleo.peercommunityin.org/articles/rec?i…


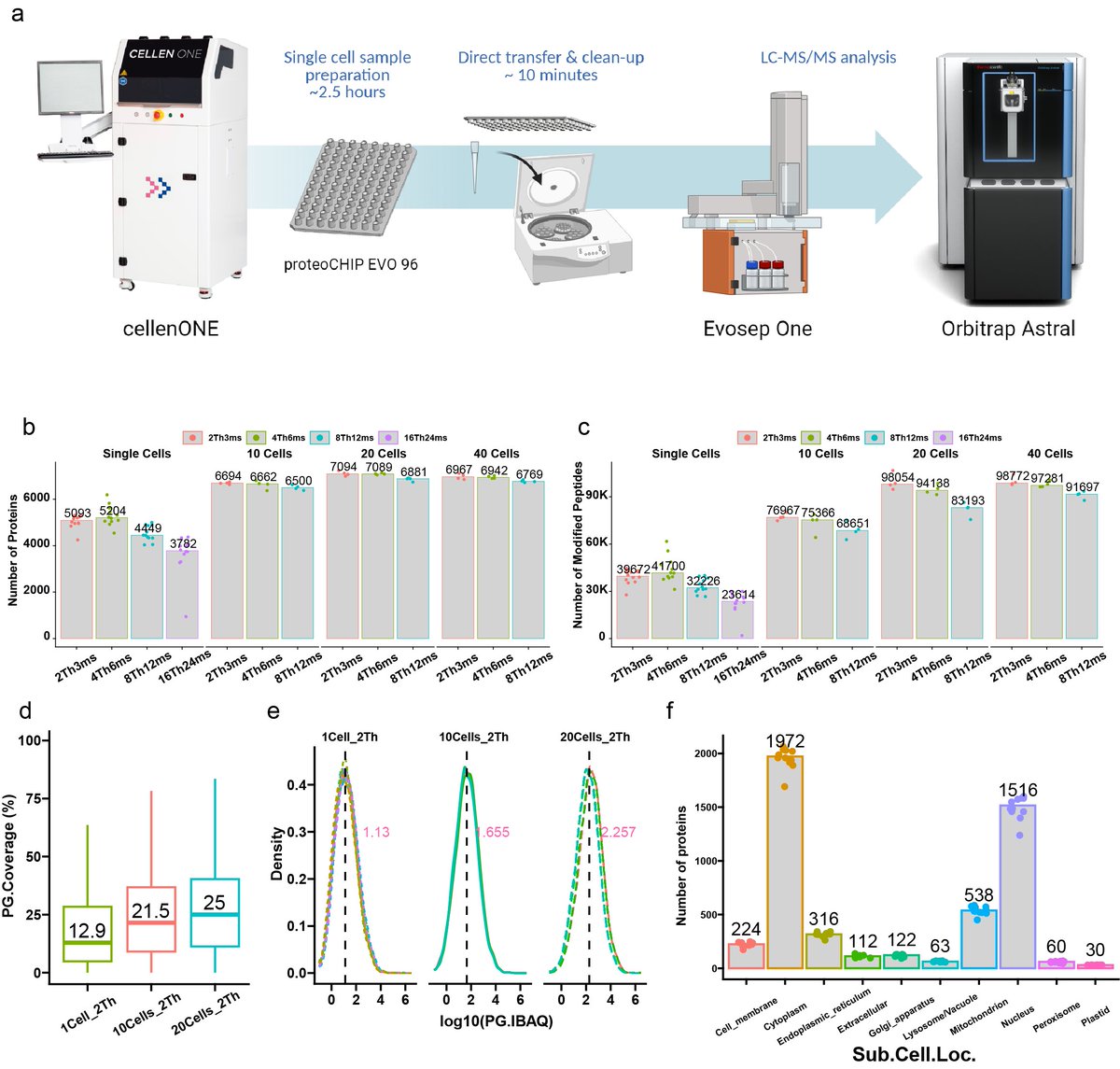
Excited to share our latest research on thorough optimisation of phosphopeptide enrichment parameters that also includes Astral MS #phosphoproteomics data ✨ Thanks to Ana Martinez del Val, @BortelPatricia, Ilaria Piga and Claire Koenig. biorxiv.org/content/10.110…



💀🔬 Exciting development in #palapeoproteomics! Streamlined MS workflow for biological sex identification from 🦷 developped by Claire Koenig, making confident and rapid sex determination accessible for large-scale studies. #proteomics #TeamMassSpec biorxiv.org/content/10.110…

Curious about #phosphoproteomics? Check out our latest paper deciphering phosphopeptide enrichment and showcasing sensitivity on both Exploris and Astral MS ✨. Great work @BortelPatricia, Ana Martinez del Val, Ilaria Piga and Claire Koenig. #TeamMassSpec mcponline.org/article/S1535-…

Interested in obtaining the ultimate analytical depth of the phosphoproteome? Please join Stoyan Stoychev hosting @BortelPatricia for episode 18 of our EMERGE webinar series on 24 April! Showcasing latest innovative strategies with JesperOlsenLab #Astral us06web.zoom.us/webinar/regist…


📢 Join us tomorrow for the next talk in our #PAASTA Seminar Series which will be given by Claire Koenig!!












#HumanEvolution discovery! Our new study in Science Magazine uses #paleoproteomics to reveal the biological sex and genetic diversity of 2-million-year-old #Paranthropus robustus from South Africa! Read more: science.org/doi/10.1126/sc… #Paleoanthropology #AncientProteins