clement cazorla
@cazorla31
I am a postdoc researcher in bioimage analysis at CBI Toulouse. Main author of Svetlana and Sketchpose Napari plugin 😎
ID: 1722246143932006400
https://koopa31.github.io/ 08-11-2023 13:34:32
47 Tweet
48 Followers
84 Following


Hello there ! I am now part of Julie Batut’s team at CBI (Toulouse) as a postdoctoral researcher to work on olfactive cells tracking in zebra fish embryos on 3D spinning disk microscope images ! Stay tuned 😎 cbi-toulouse.fr/fr/equipe-batut





So happy to see how far napari has gone! The repo was created with Juan Nunez-Iglesias on the Caltrain on the way to the Computer History Museum Museum. Fast forward 6 (!) years and napari is an essential tool for bioimaging and the Python Science ecosystem. Thanks to a thriving


If you're not already following Christopher Hamblin, you definitely should! 😉 I'm super proud of this work, and there's more on the way. In case you're curious, Chris created these images without diffusion—just pure gradient ascent on a classifier. A true magician 🐇


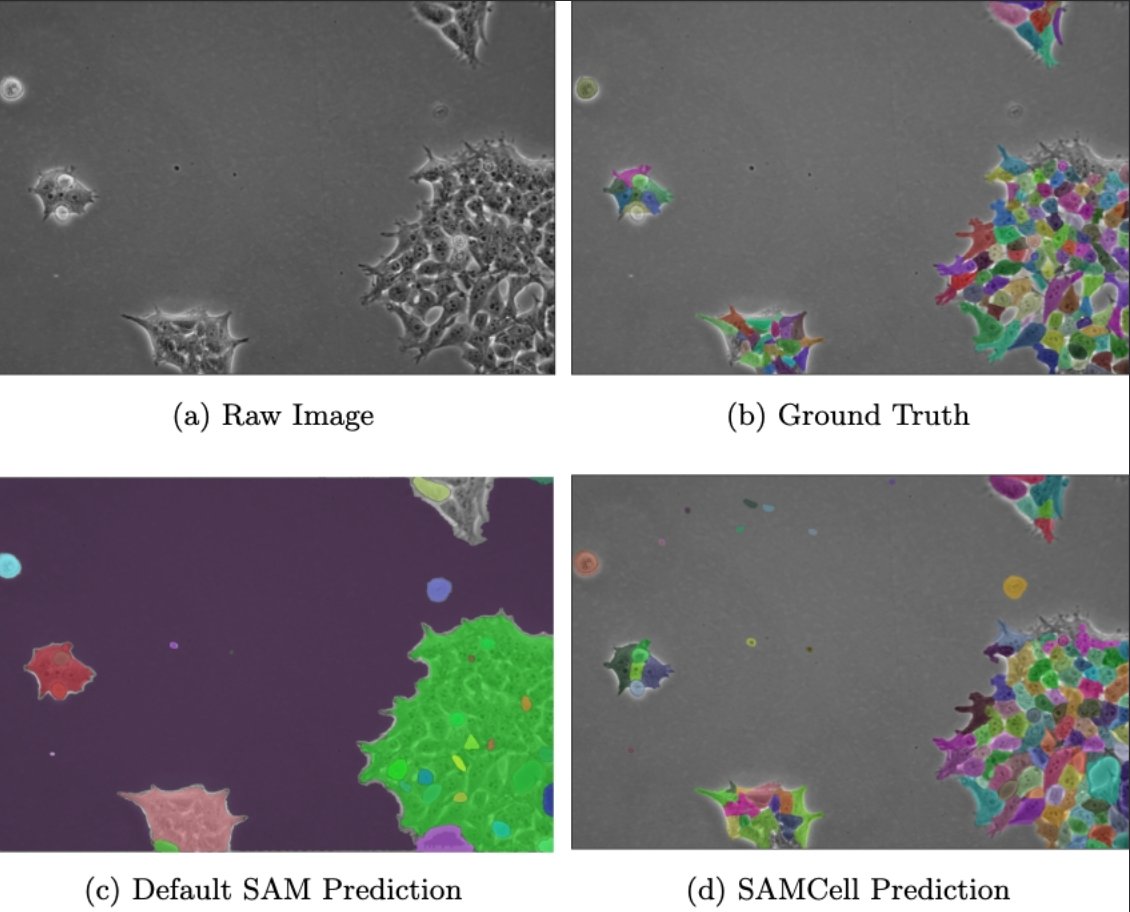
Researchers at IMACTIV-3D and Les Maths à Toulouse aim to streamline the classification of #histopathology segmentation results by minimizing the amount of manual annotation. 🖊️ Learn about the plugin they've created to address this issue in Scientific Reports: go.nature.com/3VCYnKE







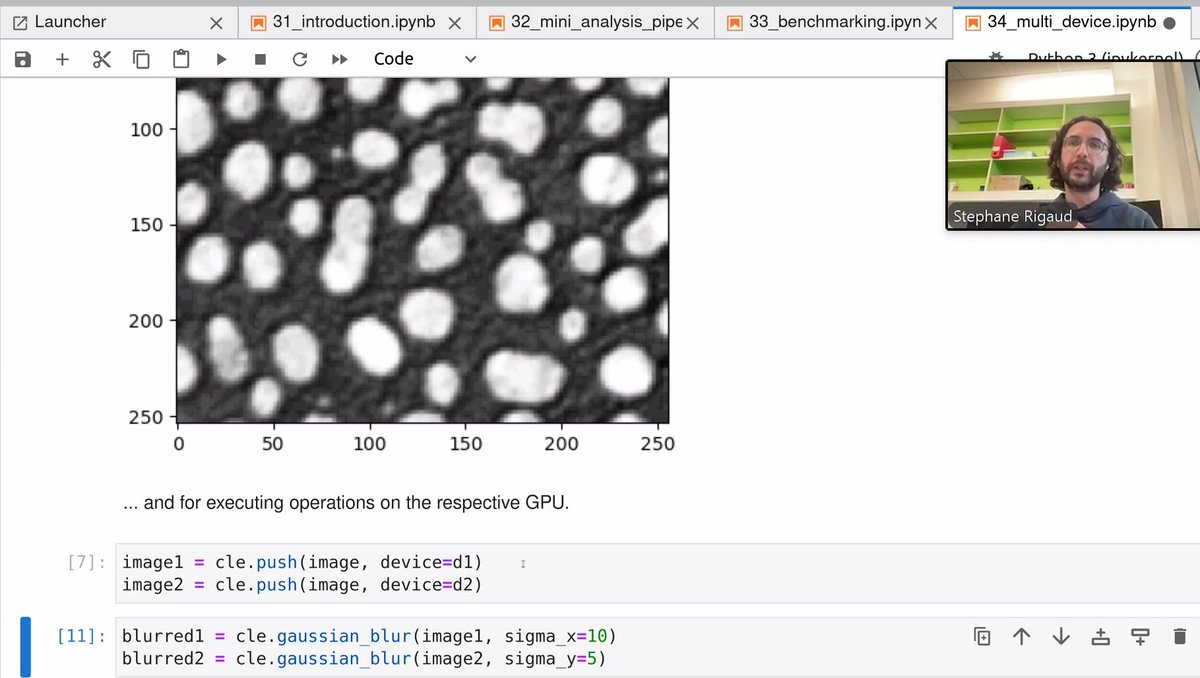
Amazing, Stephane Rigaud is demonstrating multi-GPU support🎉 using #pyclesperanto(without _prototype) for #BioImageAnalysis at #I2K2024(virtual)!🔬🖥️🚀 The notebook is available online #openaccess github.com/StRigaud/clesp…




Thrilled to share MyoFuse, an AI-based workflow for automated skeletal muscle cell fusion quantification! 🥳 This is a collaboration with Benjamin Lair and Cedric Moro (I2MC-Toulouse), improving the Fusion Index (FI) measurement. Preprint: biorxiv.org/content/10.110…