Carlos Alvarez
@carlos_alvarezq
PhD candidate at Centre for Synthetic Biology and enzyme hunter looking for his next target
ID: 1322488266923380737
31-10-2020 10:39:24
691 Tweet
141 Followers
399 Following

Sharing our press release on the significance of our recent Nature Communications article, along with my personal perspective on its impact. Hope you enjoy the read! 🔗 pan.pl/en/ippt-pas-sc… Thoughts and feedback welcome!


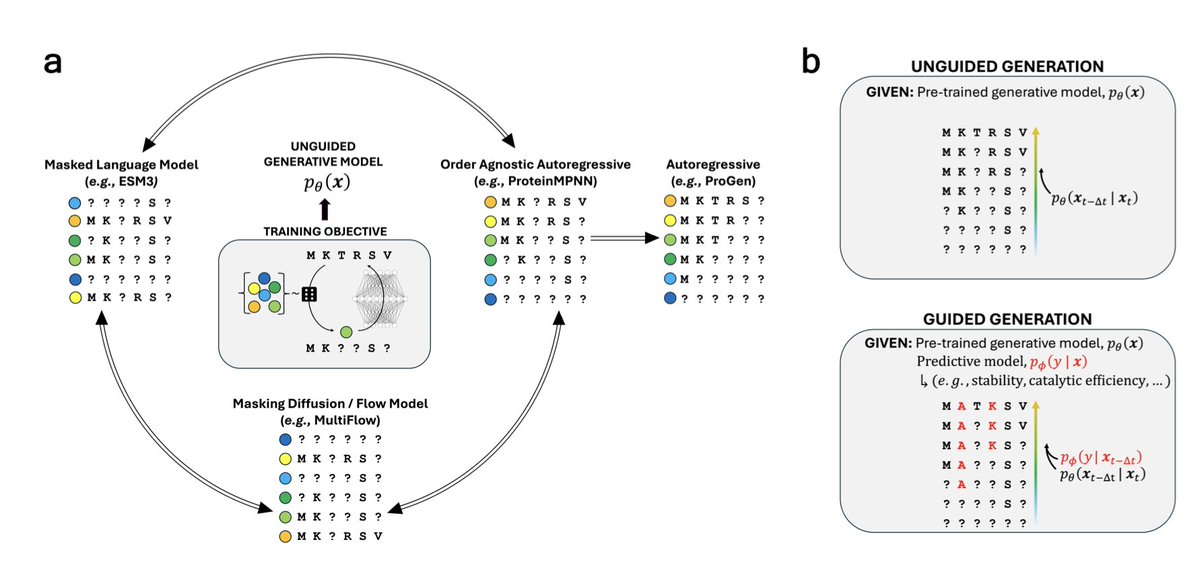
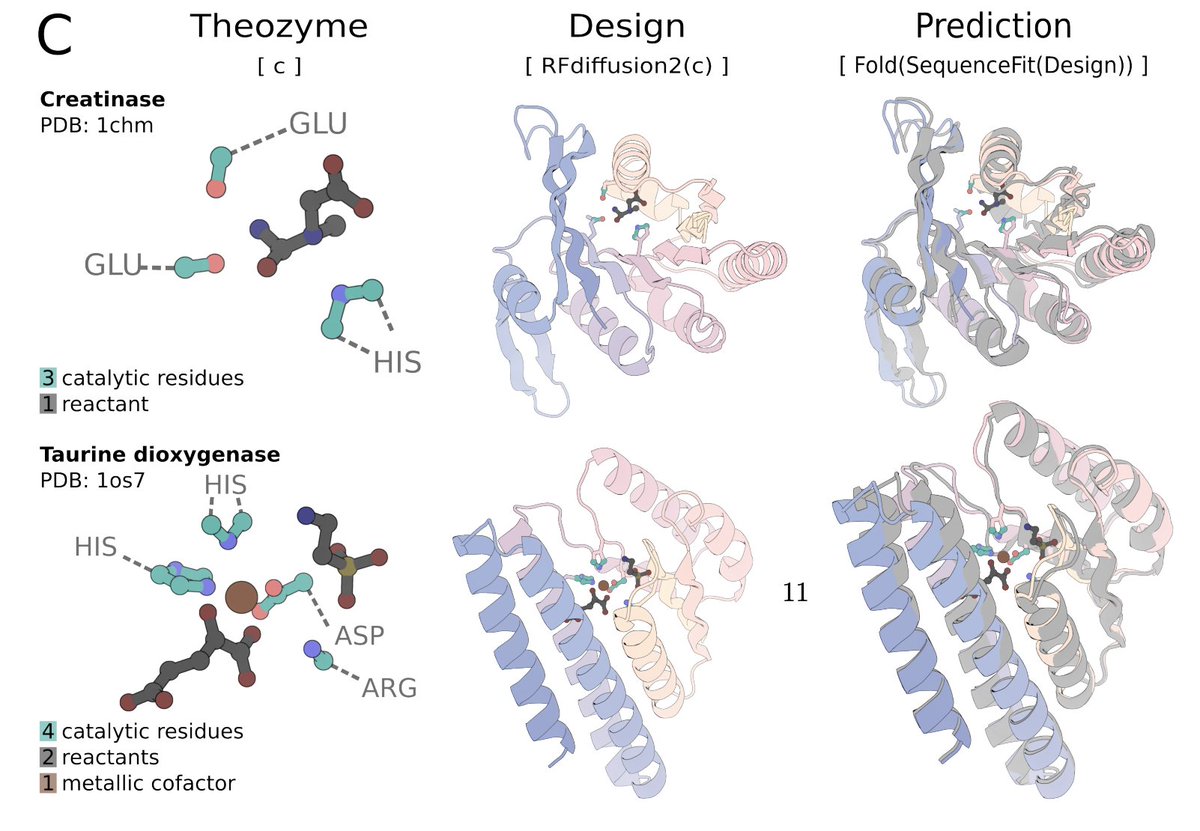
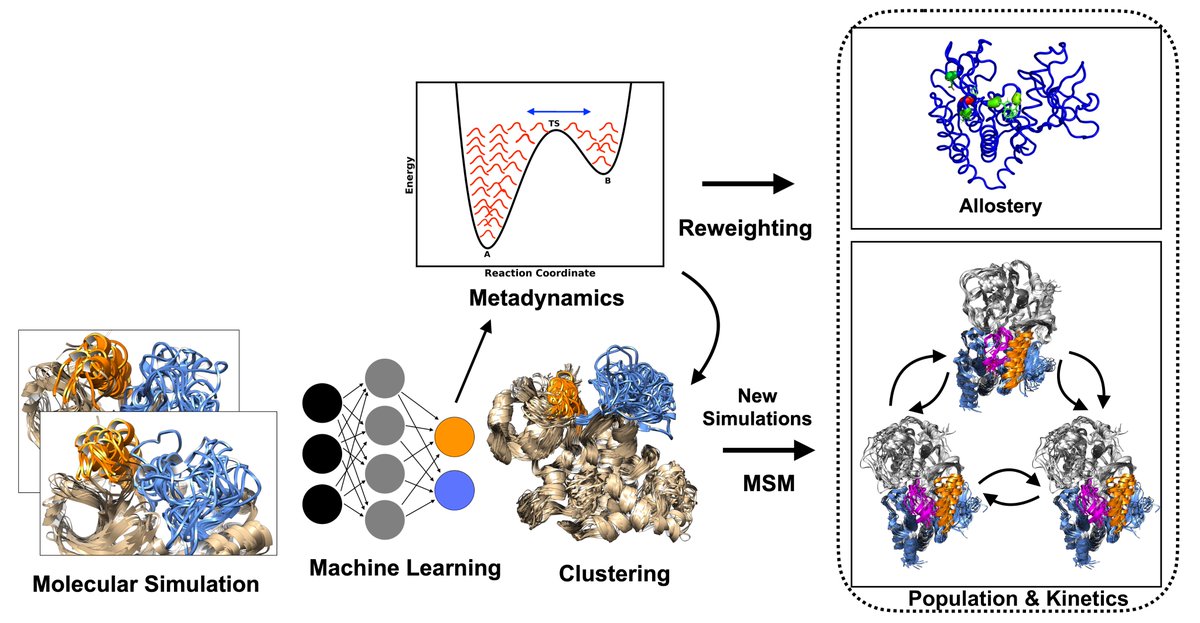
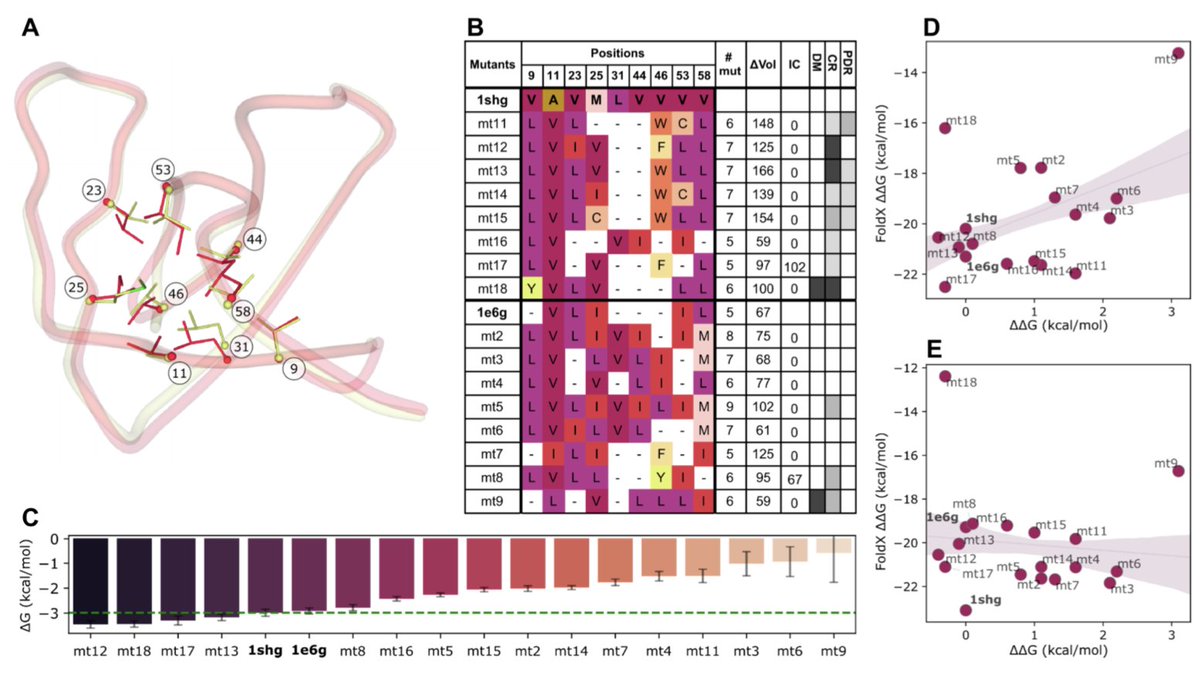
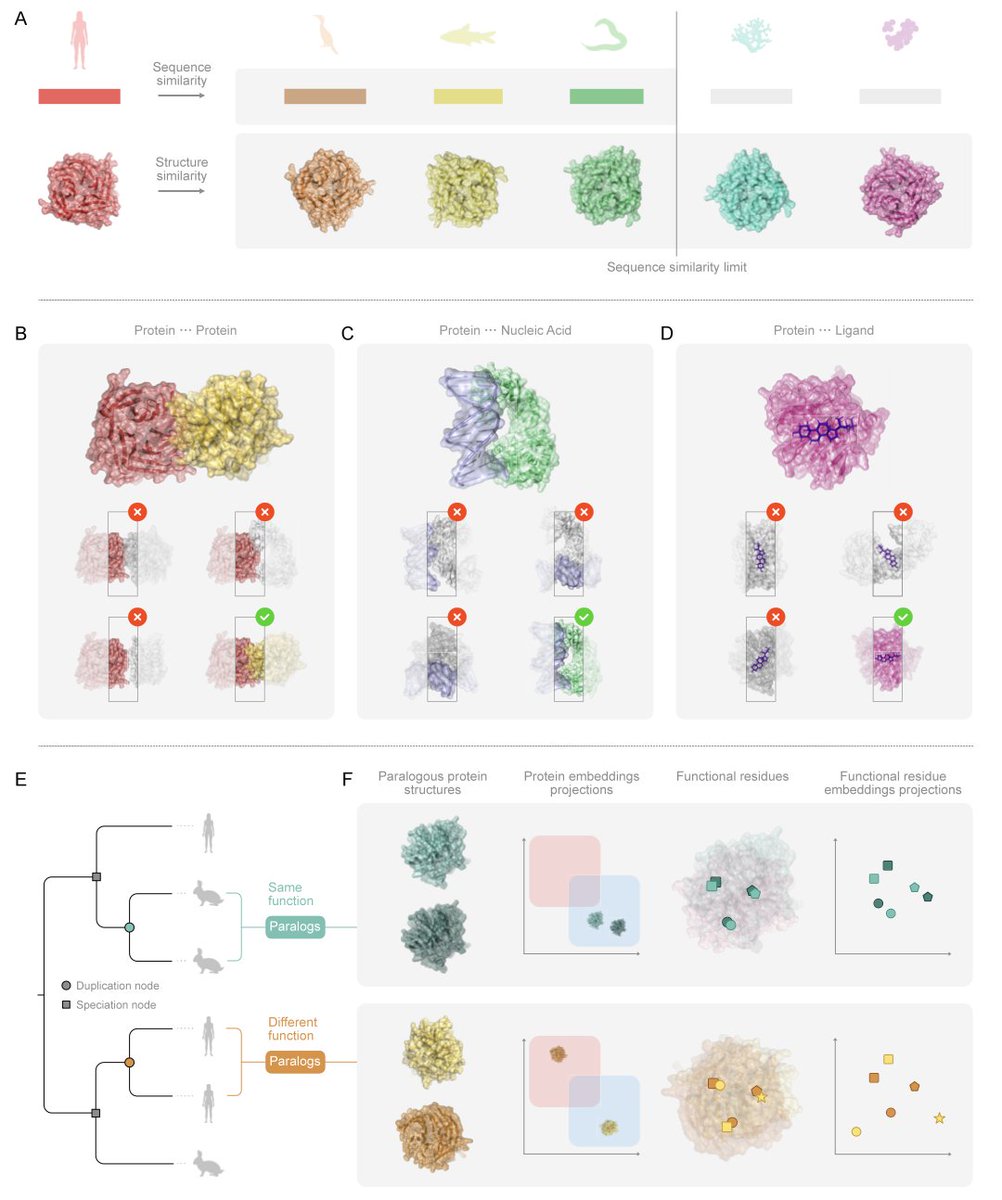
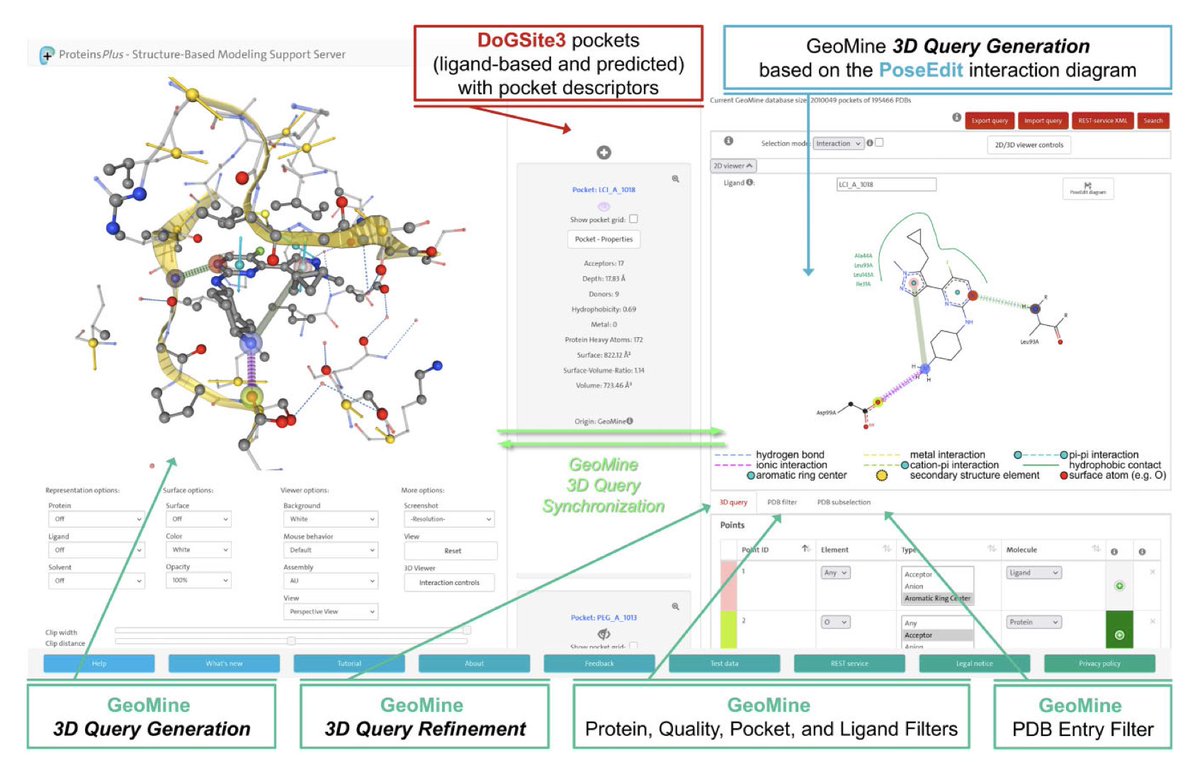
ProteinsPlus: a publicly available resource for protein structure mining Nucleic Acids Res 1. ProteinsPlus is an open-access web server offering a comprehensive suite of tools for protein structure analysis, protein-ligand modeling, and PDB mining—making it one of the most versatile


🚨 Major Update: We’ve released ColabSeprot, the sister of ColabSaprot, featuring ESM1b, ESM2, ProTrek, ProtBert! 🎉 Fine-tune protein language models in just a few clicks—no coding required! 🧬✨ 🔗 ColabSeprot: colab.research.google.com/github/westlak… ColabPLM video: youtube.com/watch?v=nmLtjl…



Nice study from AstraZeneca (EvaNittinger, Özge Yoluk, Alessandro Tibo, Gustav Olanders) on the prediction of allosteric vs orthosteric ligands using a curated set of 17 targets! I've always been curious about the performance of cofolding models on allosteric interactions and











🚀 Excited to release BoltzDesign1! ✨ Now with LogMD-based trajectory visualization. 🔗 Demo: rcsb.ai/ff9c2b1ee8 Feedback & collabs welcome! 🙌 🔗: GitHub: github.com/yehlincho/Bolt… 🔗: Colab: colab.research.google.com/github/yehlinc… Sergey Ovchinnikov Martin Pacesa

We just published a quick write-up on BoltzDesign1—the open-source all-atom protein design tool from Yehlin Cho, Sergey Ovchinnikov , Martin Pacesa & Bruno Correia . Congrats to the team for pushing accessible binder design forward! ariax.bio/resources/bolt…