Sebastian Prillo
@sebastianprillo
EECS PhD student, UC Berkeley
ID: 908064056048848898
13-09-2017 20:25:16
14 Tweet
31 Takipçi
43 Takip Edilen

Our newest work is out! Single Layers of Attention Suffice to Predict Protein Contacts This was a wonderful collaboration with Nick Bhattacharya, @roshan_m_rao Justas Dauparas David Baker (Institute for Protein Design) Peter Koo Yun S. Song Sergey Ovchinnikov biorxiv.org/content/10.110… 🧵 1/n

We have ≥$10k to support talented 14-18 year olds whose studies were interrupted by war in Ukraine. We especially would like to hear from IMO, EGMO, MEMO, IOI, EGOI, IPhO, IChO contestants. If you're one or know one, here's the form to apply: ferenchuszar1.typeform.com/ukrainefund-eng

A Stanford University professor just threatened me with police. After BBQ Becky, Permit Patty, Golfcart Gail, and all the memes, we now have Retweet Rachel. Public advisory: don't call the cops on black people for no reason. Black people disagreeing with you on Twitter is not a crime.



Very glad to see our CherryML method published online in Nature Methods. The published version has updated and expanded results, so please check it out. doi.org/10.1038/s41592… (1/3)






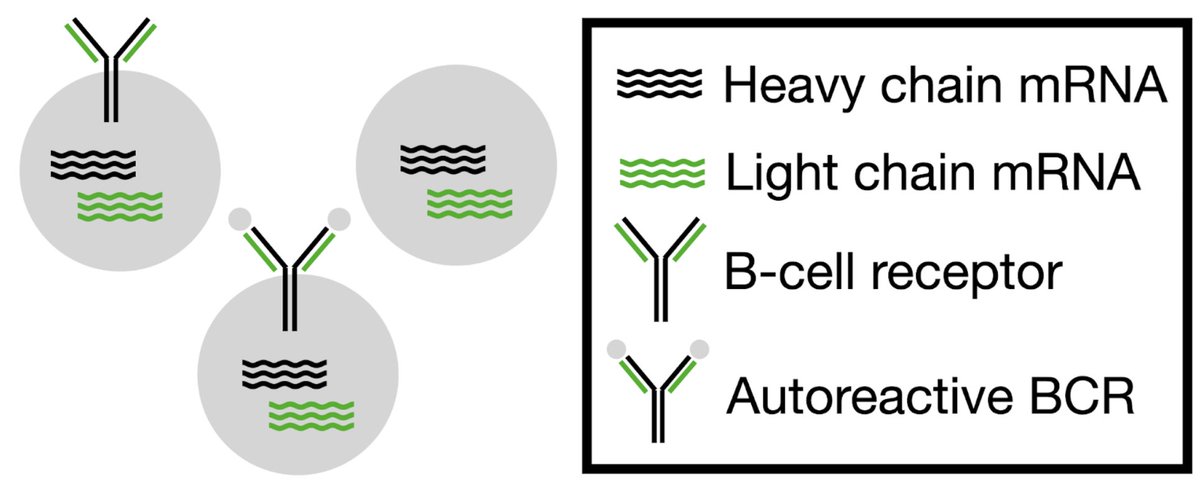
Antibodies are highly diverse, but most possible sequences are unstable or polyreactive. In this work led by Milind Jagota, we propose a new source of data for modeling constraints from these properties. Our models show clear improvements in predicting antibody dysfunction.(1/n)

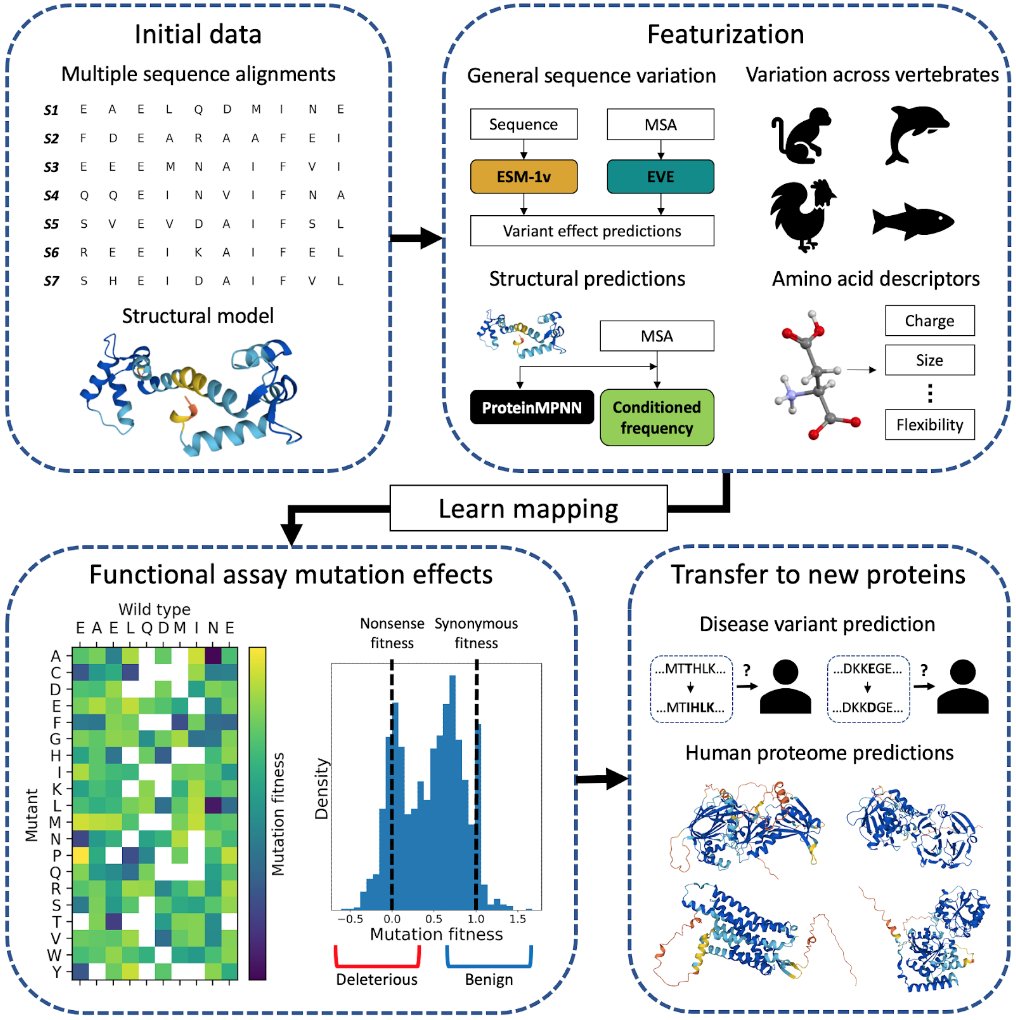
Large protein language models can learn complex epistatic interactions, but how much does that help with predicting variant effects? In this NeurIPS article, we show that classical independent-sites phylogenetic models can outperform pLMs on this task. 1/2 openreview.net/forum?id=H7mEN…


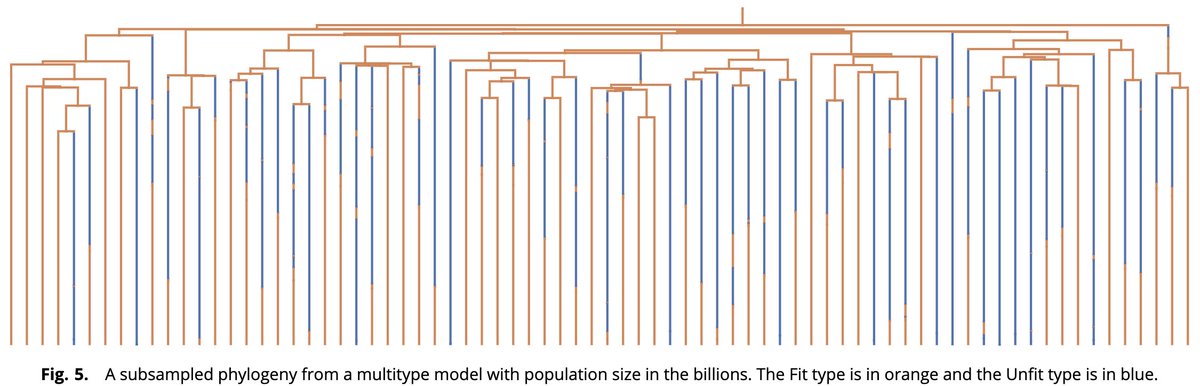
How can one efficiently simulate phylodynamics for populations with billions of individuals, as is typical in many applications, e.g., viral evolution and cancer genomics? In this work with Michael Celentano, W. DeWitt, & S. Prillo, we provide a solution. doi.org/10.1073/pnas.2… 1/n