
Matthew Lewis
@mreeselewis
ID: 747419144908324864
27-06-2016 13:19:45
19 Tweet
86 Takipçi
62 Takip Edilen



"Learning and doing lab work in a friendly and supportive environment, which I found at the NationalPhenomeC(UK), was extremely important for my own advancement in research." Hear about Ziyue's experiences on our MRes in Biomedical Research imperial.ac.uk/metabolism-dig…



We're always looking to improve the platform, innovate and get more from our existing data here NationalPhenomeC(UK), read about our new thoughts on #NMR in our paper out today! #metabolomics doi.org/10.1039/D0SC01…



Very proud of Caroline Sands and the NationalPhenomeC(UK) team for a fantastic effort - shining light on & significantly advancing QC in LC-MS metabolomics!

Researchers at Imperial College London provide a fresh perspective on the hidden HiFi metabolome and how to find it in your LC-MS data. NationalPhenomeC(UK) pubs.acs.org/doi/10.1021/ac…

As Partner in Medical Research Council funded MAP/UK consortium we're pleased to announce registration is open for Road/MAPUK meeting. We want end-users of #metabolomics #lipidomics to tell us what your needs are to ensure we're developing those capabilities for future mapuk.org/RoadMAP



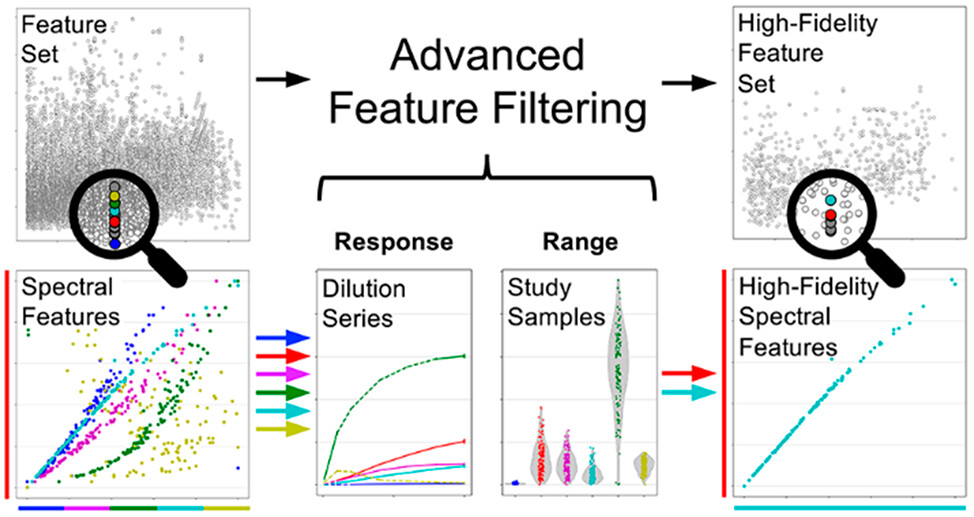
Nicely done, a thorough #metabolomics ACS Analytical Chemistry Journals effort identifying ubiquitous small peptides in urine samples.

Now online in Med by Cell Press. ID ACF Ravi Mehta found ddhC, free base of the only known antiviral small molecule in patients with COVID-19 and other viruses with input of NIHR Imperial BRC NationalPhenomeC(UK) Myrsini Kaforou Dr Ho Kwong Li Mahdad Noursadeghi Graham Cooke Cell Press cell.com/med/fulltext/S…

🧵NationalPhenomeC(UK) is excited to make available its complete “open platform” for LC-MS-based metabolomics. Read about the methods, protocols, open-source software and metabolite annotations in our new preprint on ChemRxiv doi.org/10.26434/chemr… #OpenAccess #OpenScience #Metabolomics





