
Karthik Sudarsanam
@karthick_sudars
PhD student @iitgncompbio group, @iitgn | Master's degree in Bioinformatics | Gravitated by protein structures and dynamics
ID: 1099522877554974720
24-02-2019 04:14:16
130 Tweet
83 Takipçi
483 Takip Edilen

A diffusion model for programmable protein structure generation, with code and wet-lab experiments. Andrew Beam Wujie Wang Generate:Biomedicines nature.com/articles/s4158…


My first paper in the Alberto Perez group! Opportunities, strategies, and challenges in peptide-based drug discovery wires.onlinelibrary.wiley.com/doi/10.1002/wc… Bright future ahead for the marriage between physics-based and AI models #compchem #drugdesign #MachineLearning UF Chemistry

A new version of AlphaPulldown by DingquanYu. Includes experimental support for multimeric templates in AlphaFold by Dmitry Molodenskiy and interface to crosslink-based modeling AlphaLink2 of Rappsilber laboratory github.com/KosinskiLab/Al…
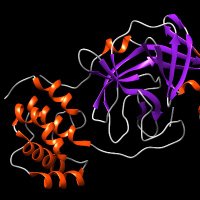


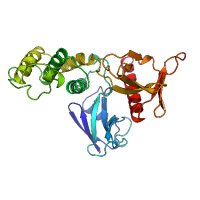




We've got a cool paper out with Grzegorz Grabe and Sophie Helaine lab! This bacterial🦠toxin-antitoxin system is doing some crazy stuff, and we could help explain the structural role of DNA in the whole process: nature.com/articles/s4159… Check out the long explainer movie I made



As Computational Biology @IITGN group starts its exploration of modeling plant circadian rhythms, some interesting results soon. In the mean time, check out a comprehensive review of differential equation based models in this area by PhD student Shashank Kumar Singh doi.org/10.1007/s11103…











