Biotite
@biotite_python
This Python package is your Swiss army knife for bioinformatics. It allows analysis of sequence and biomolecular structure data in an easy and efficient manner.
ID: 933981641260847106
https://www.biotite-python.org/ 24-11-2017 08:52:30
117 Tweet
336 Takipçi
6 Takip Edilen




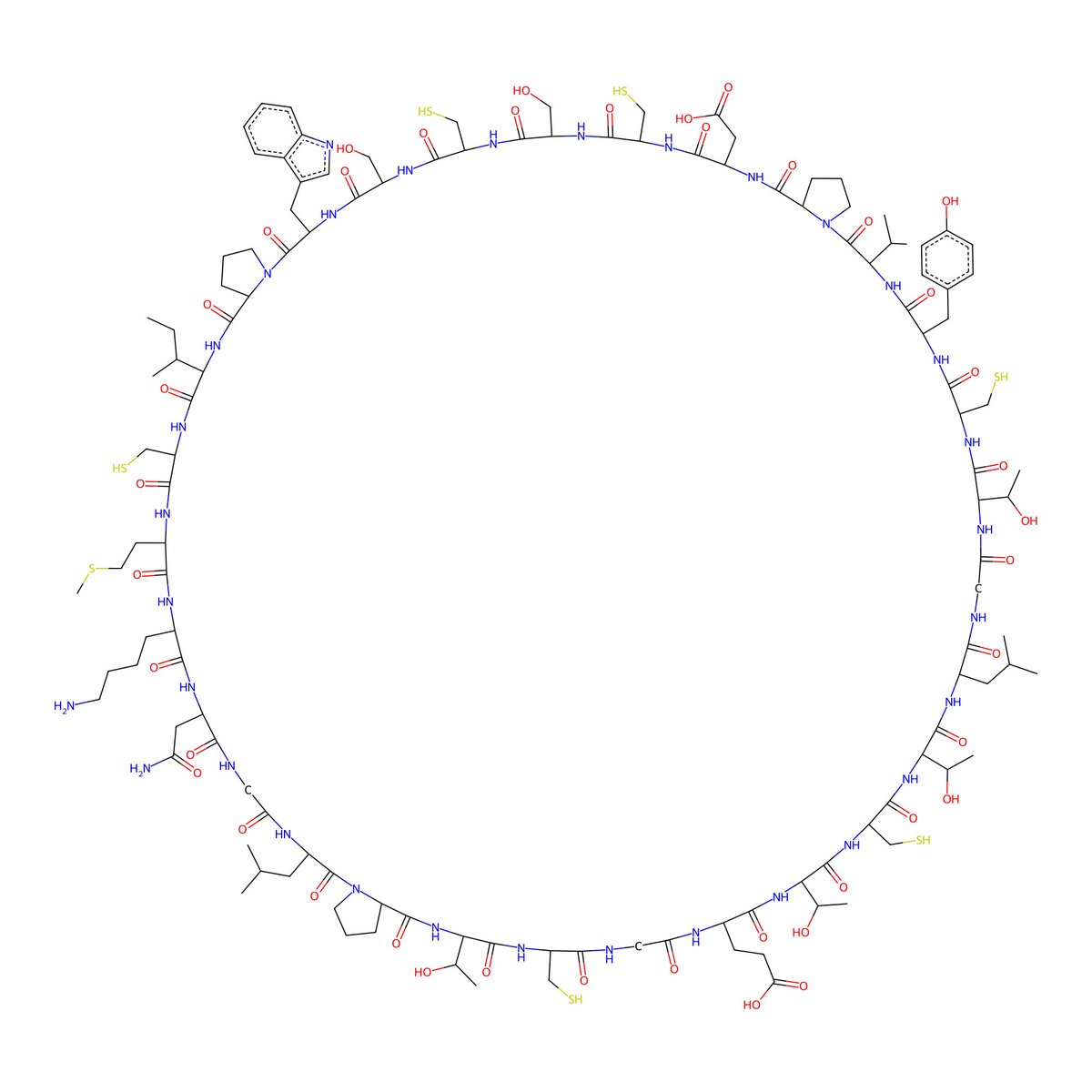
Diving into the Chemical Component Dictionary can be quite entertaining. Thank you, rcsb pdb 💉🧬💻🔬💊🌱🧠🦠. Me: Can we have 3UQ? Biotite: We have 3UQ in the PDB. 3UQ in the PDB (rcsb.org/ligand/3UQ):



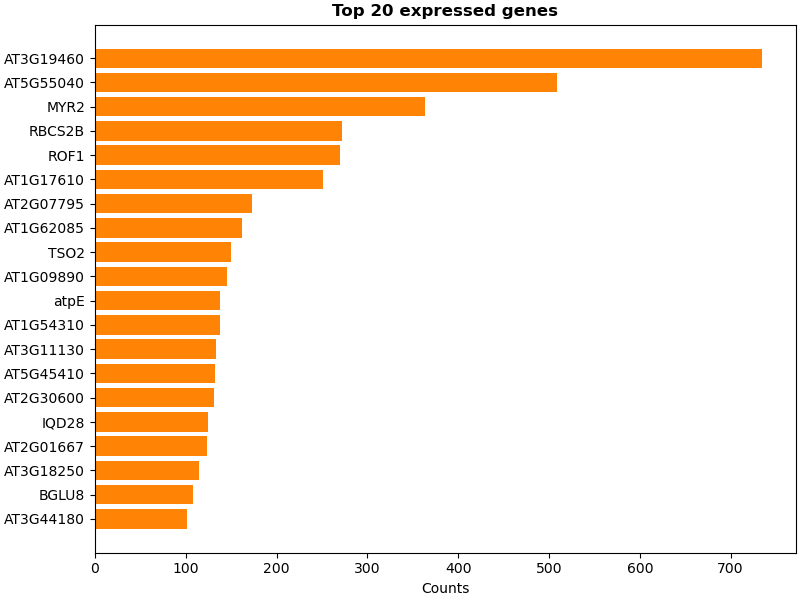
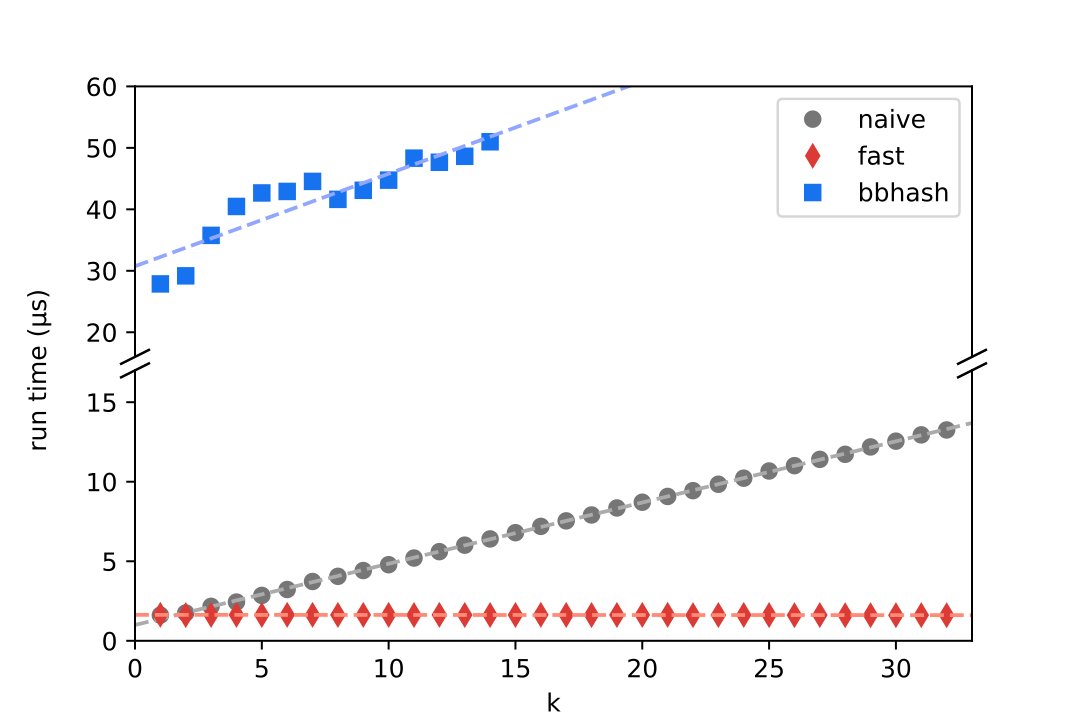
Biotite 0.41.0 has been released and it brings quite a bunch of improvements: full-fletched SDF file format, reading/writing bonds in rcsb pdb 💉🧬💻🔬💊🌱🧠🦠 NextGen PDBx files and more robust structure superimposition. Read the full changelog at github.com/biotite-dev/bi…. #Bioinformatics