Allie Greaney
@alliegreaney
MSTP MD/PhD student studying virus evolution and immunity in the @jbloom_lab at @fredhutch @uwgenome @UW_MSTP. she/her
ID: 1234267113549754368
02-03-2020 00:00:07
84 Tweet
907 Takipçi
461 Takip Edilen

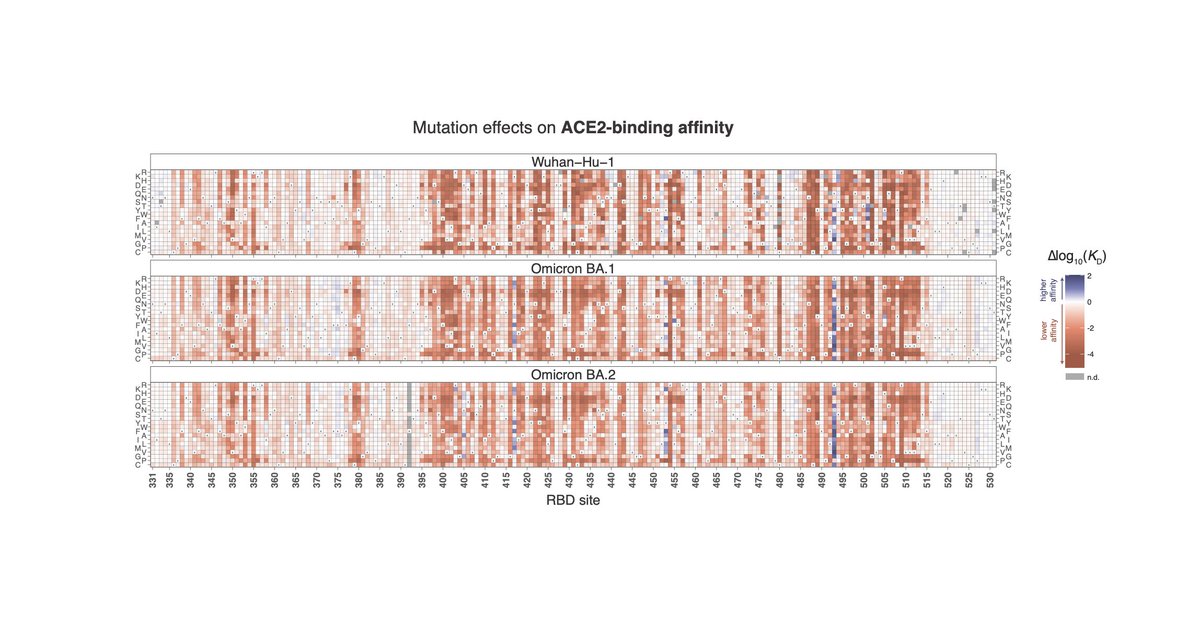
Cool graphic from nat is on bluesky now vividly illustrating potential for continuing evolution in #SARSCoV2 receptor-binding domain (RBD), which is key target of antibodies. There are 15 RBD mutations in Omicron (BA.1), which seems like a lot! But... (1/3)

Wanted to add to @DrNeeltje's nice summary about the recent pre-print on Alexander Cohen The Bjorkman Lab's mosaic RBD nanoparticle vaccine, which at least in animals induces "broader" antibody immunity to #SARSCoV2 variants and other SARS-related CoVs: biorxiv.org/content/10.110… (1/n)

Our work on the stabilization of flu NA is officially published in Nature Communications! Large congrats and thanks to Julia Lederhofer, @kanekiyom, King Lab, The Veesler Lab, Oliver Acton, Barney Graham, MD, PhD and many more

What a special moment: the #COVID19 vaccine we designed with Lexi Walls King Lab has met its primary endpoints in the phase 3 clinical trial run by SK bioscience!!! HHMI Institute for Protein Design UW Biochemistry 1/4 newsroom.uw.edu/news/uw-medici…

Excited to share our work on epistasis shaping the evolution of SARS-CoV-2 BA.1 — had a blast working on this with Alief Moulana @ThomasDupic Jeffrey Chang, Michael Desai & Bloom Lab: biorxiv.org/content/10.110…

If you’re into viral, antibody, or protein evolution, check out the 🌟 Starr lab 🌟! Can confirm that Tyler Starr is one of the most brilliant, fun, and supportive scientists out there! Already envious of the new folks who get to work with you!

We have a position in the Bloom lab for a research tech interested in using deep mutational scanning to study evolution of viruses including SARS-CoV-2 & influenza: careers-fhcrc.icims.com/jobs/22925/res… (And while this ad is for a tech, we are always open to inquiries from postdoc candidates)

Our latest review is now out in Biochemistry! In this Perspective, we discuss the importance of InDels in protein evolution, whether in nature or the test tube, and strongly advocate their potential for protein engineering. Tokuriki lab pubs.acs.org/doi/10.1021/ac…

In new study led by David Bacsik, we show single influenza-infected cells produce wildly different numbers of viral progeny: biorxiv.org/content/10.110… Surprisingly, cells that transcribe the most viral mRNA are NOT ones that produce the most progeny virions. Explanatory 🧵 below


Was a pleasure to contribute to this very cool study looking at how heterologous boosting elicits new versus boosted original antigenic sin antibody responses in mice from Victora Lab !



In work led by @bdadonaite Kate H D Crawford Caelan Radford, we develop pseudovirus system to deep mutational scan full #SARSCoV2 spike: biorxiv.org/content/10.110… TLDR: can now safely measure how all mutations to viral entry proteins affect antibody neutralization. 🧵 below (1/n)