Ahmet Rifaioğlu
@ahmet_rifaioglu
Researcher in @UniHeidelberg @uniklinik_hd | PhD & MSc @METU_ODTU
ID: 1198229096
19-02-2013 18:18:02
263 Tweet
2,2K Followers
420 Following


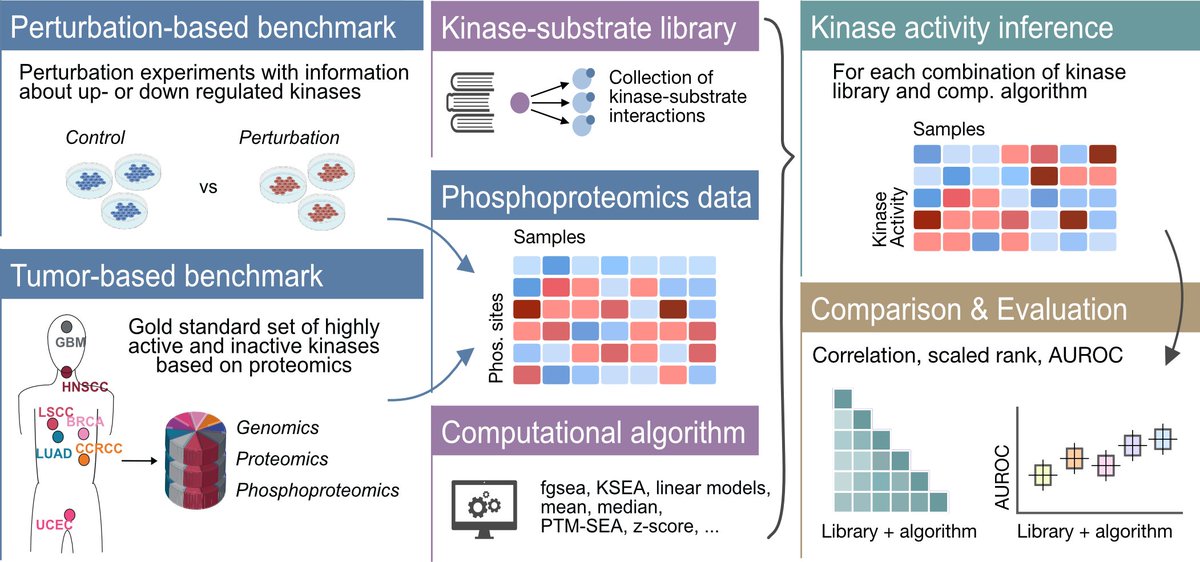
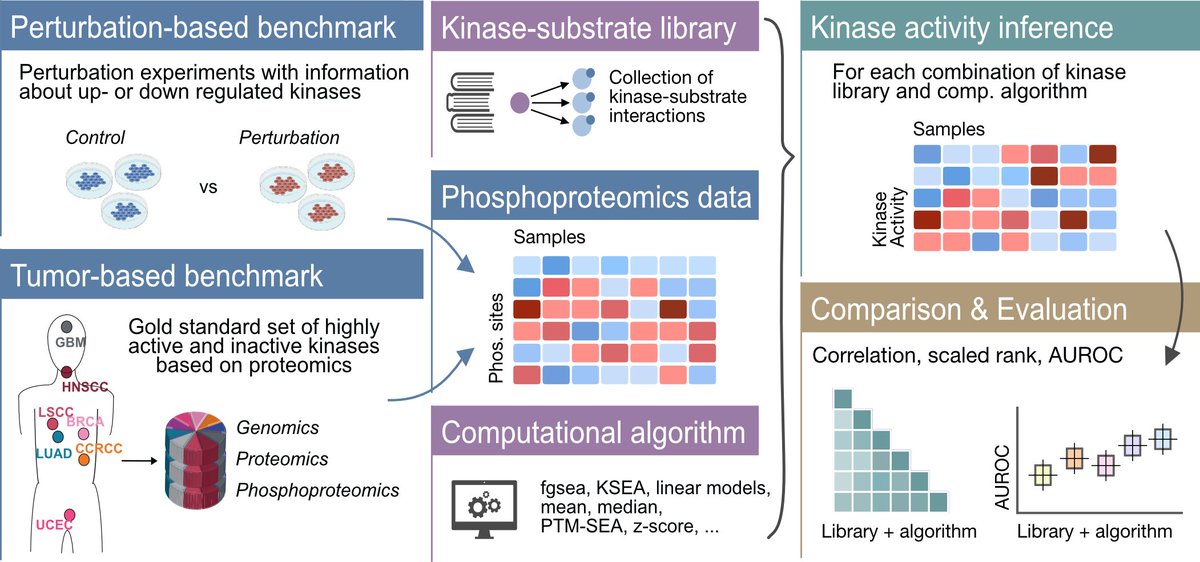
Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭In collaboration with the Zhang lab BCMHouston, we performed a comprehensive evaluation of kinase activity inference tools to help answer that question tinyurl.com/yc56axsv 🧵⬇️



📣 The much-revised manuscript of LIANA+, our all-in-one solution to study cell-cell communication from single-cell, spatial, and multi-omics technologies, is now published in Nature Cell Biology nature.com/articles/s4155…


Post-doc opening to join our lab EMBL-EBI #EMBLEBIresearch to develop/apply #bioinformatics methods to study cellular organization, int/extra cell networks to extract disease mechanisms from sc and spatial multiomic data, with Yvan Saeys, supp by Chan Zuckerberg Initiative 👇embl.org/jobs/position/…


Our work with Lucas Schirmer in multiple sclerosis (MS) is out today in Nature Neuroscience. We investigated lesion progression and cell-cell communication events in MS using snRNA-seq and spatial transcriptomics 🧠 See below ⬇️ nature.com/articles/s4159…



We are excited to announce an opening for a PhD student to join our research team at Uniklinik Heidelberg. If you are passionate about unraveling the molecular mechanisms underlying metabolic disorders, please apply at: karriere.klinikum.uni-heidelberg.de/index.php?ac=j… Deutsches Zentrum für Diabetesforschung (DZD) Helmholtz Diabetes Center (HDC)

🚀"Signals and Systems: Theory and Practical Explorations with Python" by our faculty members Fatos Yarman Vural (Fatos Tunay Yarman Vural) and Emre Akbas (Emre Akbas) is out from Wiley! Learn more at wiley.com/en-us/Signals+…



🧠 Excited to share my main PhD project! We mapped the regulatory rules governing Glioblastoma plasticity using single-cell multi-omics and deep learning. This work is part of a two-paper series with Omer Ali Bayraktar , Oliver Stegle and Moritz Mall groups. Preprint at end🧵👇
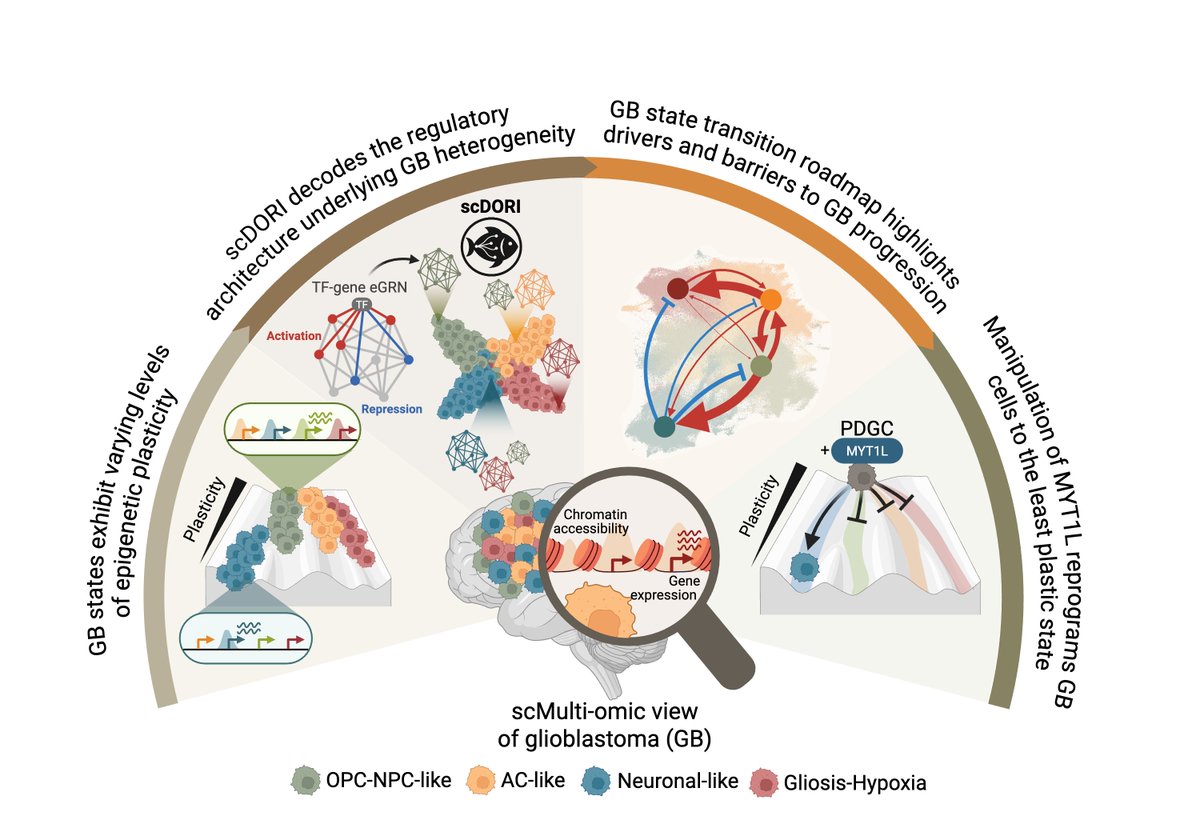

Great effort by Omer Ali Bayraktar, Stegle Lab & Moritz Mall labs in unravelling GBM cell state variability and spatial organisation at single-cell multimodal resolution. Happy that we could also contribute to this fantastic study! #Glioblastoma #SingleCell #SpatialOmics

Great work by Omer Ali Bayraktar , Oliver Stegle , and Moritz Mall ! Happy that our Ahmet Rifaioğlu contributed to this study by mapping CCC events between spatially co-localized TME and malignant cells using LIANA+ 🧠 github.com/saezlab/liana-…

Ever wondered which method to use to infer kinase activities from phosphoproteomics data? 💻💭Our revised comprehensive evaluation of kinase activity inference tools, done in collaboration with the Zhang lab BCMHouston, is now out Nature Communications 🔬 tinyurl.com/4twuc6z4