Alex Graudenzi
@agraudenzi
associate professor @unimib || biomedical data science || opinions are my own
ID: 1357283390484082691
https://dcb.disco.unimib.it/people/graudenzi-alex/ 04-02-2021 11:02:38
125 Tweet
1,1K Takipçi
4,4K Takip Edilen


Nice new work by Fabrizio Angaroni and colleagues! Kinbiont is a new framework for analyzing microbial kinetics using advanced numerical methods and machine learning, with great potential to enhance our understanding of microbial behavior. Keep up the great work!

The 24 #NobelPrize in physics and chemistry are de facto prizes to #AI On Friday 15, Marco Antoniotti and the Science School of Università degli Studi di Milano-Bicocca will host a day-long symposium in which experts will explain where it all started and what to expect next Registration: sites.google.com/unimib.it/nobe…


Postdoc opportunity (18 mos) Join our team to develop pipelines for single-cell seq data in an Fondazione AIRC per la ricerca sul cancro funded project! Looking for bioinformaticians and data scientists passionate about cutting-edge research (and fun too!) Apply by Dec 3rd: unimib.it/ateneo/gare-e-… please RT!


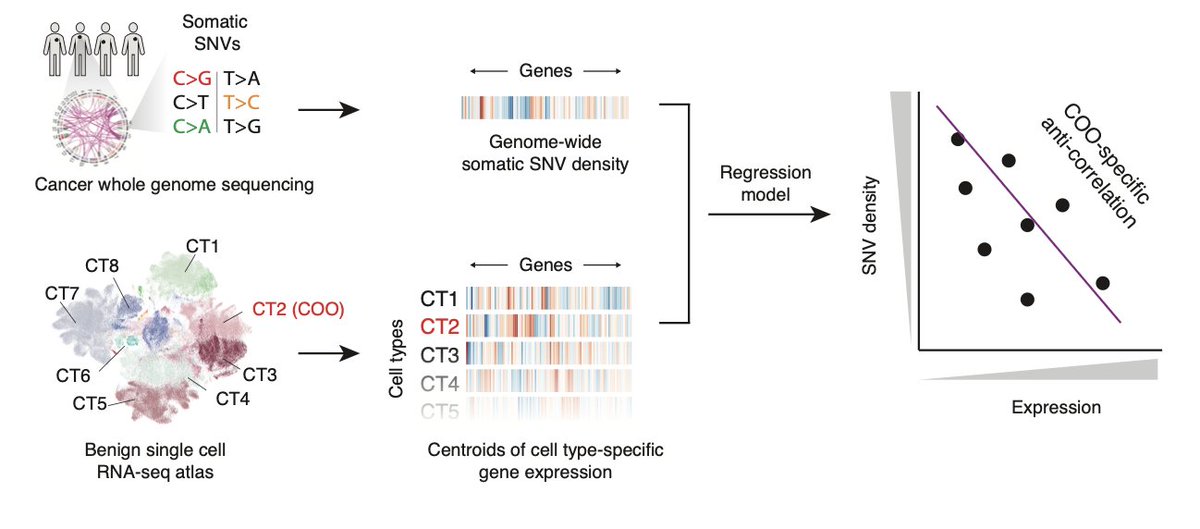
🚨So proud to present the first study from our Computational Cancer Genomics (CCG) Research Group group together with Charles Swanton Nnennaya Kanu and led by Olivia Lucas with key collaboration Sophie Ward and fundamental support Rija Zaidi Abi Bunkum & many others Out in Nature Genetics 👇 doi.org/10.1038/s41588… 🧵👇


Glad to share our newest framework spFBA allows one to estimate the fluxes of metabolic reactions from any spatial transcriptomics dataset In CRC samples metastases mimic the metabolism of the tissue of origin! Great work led by Chiara Damiani Università degli Studi di Milano-Bicocca biorxiv.org/content/10.110…


Glad to share our latest work on drug-induced tolerance in #cancer! We show that chemotherapy triggers a transient tolerant state via autophagy and DNA repair activation, which later shifts to persistence Out now in Nature Communications! doi.org/10.1038/s41467…

Glad to present the 2025 ed of the Milan Meeting on Next Generation Sequencing | Tuesday 6 May Università degli Studi di Milano-Bicocca A meeting that has become a tradition for those working on the generation or analysis of #omics data! Registration is free, coffee ☕️ too! Info at: dcb.disco.unimib.it/mmngs2025/


The Milan Meeting on Next Generation Sequencing #MMNGS 2025 is just one week away! The program is now available at this link: dcb.disco.unimib.it/mmngs2025/ 📅 6 May | Aula Martini U6 building Università degli Studi di Milano-Bicocca


🚀 EU-funded PhD in Data Science. Join us to fight AMR 🧫 at Human Technopole 🇮🇹 Milan — ENDAMR MSCA Network. 🗓 Apply by 23 May 2025 → careers.humantechnopole.it/o/phd-in-data-… 🔄 RT #PhDposition #AMR #DataScience



Very glad to be part of the team behind this work out now in Cancer Research! By combining #single_cell multi-omics data integration, #MachineLearning , and computational modeling, we map how resistance to #cancer therapies emerges and evolves doi.org/10.1158/0008-5… Università degli Studi di Milano-Bicocca


Less is more? Our new RESOLVE framework fits cancer mutational profiles with fewer #MutationalSignatures than those included in catalogues (eg COSMIC Sanger), revealing that a few key processes drive most cancers, whereas rare ones play a minor role Out now in @NAR_Open!


Incredibly proud of my former lab member Fabrizio Angaroni and his work with Fernanda Pinheiro's team on this great new paper, out now in Nature Communications! The #compbio community working on #bacteria should give it a read!


latest from the lab --> how we used whole genome passenger somatic mutation patterns 🧬 to trace the origins of never-smoker lung adenocarcinoma (LUAD) to cells of the proximal lung 🫁 Nature Genetics nature.com/articles/s4158… 1/🧵