
Adrien Leger
@adrienleger2
Director of Modified Bases Research at @nanopore. Moving slowly to where the sky is less depressing. bsky.app/profile/adrien…
ID: 2812666794
16-09-2014 08:22:36
2,2K Tweet
867 Followers
235 Following




Low coverage (5x) Oxford Nanopore data vs methylation arrays delivers far more data. Great talk by the wasatch team at #ashg2024.



New blog: Modified Base Best Practices & Benchmarking for Oxford Nanopore 🧬 sequencing Includes: * Raw POD5 files for synthetic ground truth strands * Modified base calling best practices * Modkit recommendations Check it out: labs.epi2me.io/mod-validation… #methylation #bioinformatics

It's Christmas in advance with a very special Oxford Nanopore #opendata release ! 🎅 Today we are releasing our 5mC 5hmC and 6mA DNA mod synthetic control datasets with each mods in all possible 5 mers contexts. All nicely wrapped in a comprehensive validation blog post.🎁


I am absolutely thrilled to be back in Kanazawa, Japan (a fave!), for the #tRNA2024 conference! It was my pleasure to present recent updates for Oxford Nanopore #directRNA sequencing, including our ever-expanding mod base seq offerings to this very keen audience! #tRNA #RNAmods


The latest hifiasm can directly assemble standard Oxford Nanopore simplex R10 reads, without HERRO correction or other preprocessing, to phased contigs of contiguity comparable to HiFi assembly. Like before, you can further add ultra-long, Hi-C or trio data for better assembly.

Excited about Oxford Nanopore's collaboration with UK Biobank to create the first comprehensive epigenetic map of the human genome. Thrilled to see work from our team (Marcus Stoiber, Adrien Leger, ArtRand ) driving such pivotal advancements!

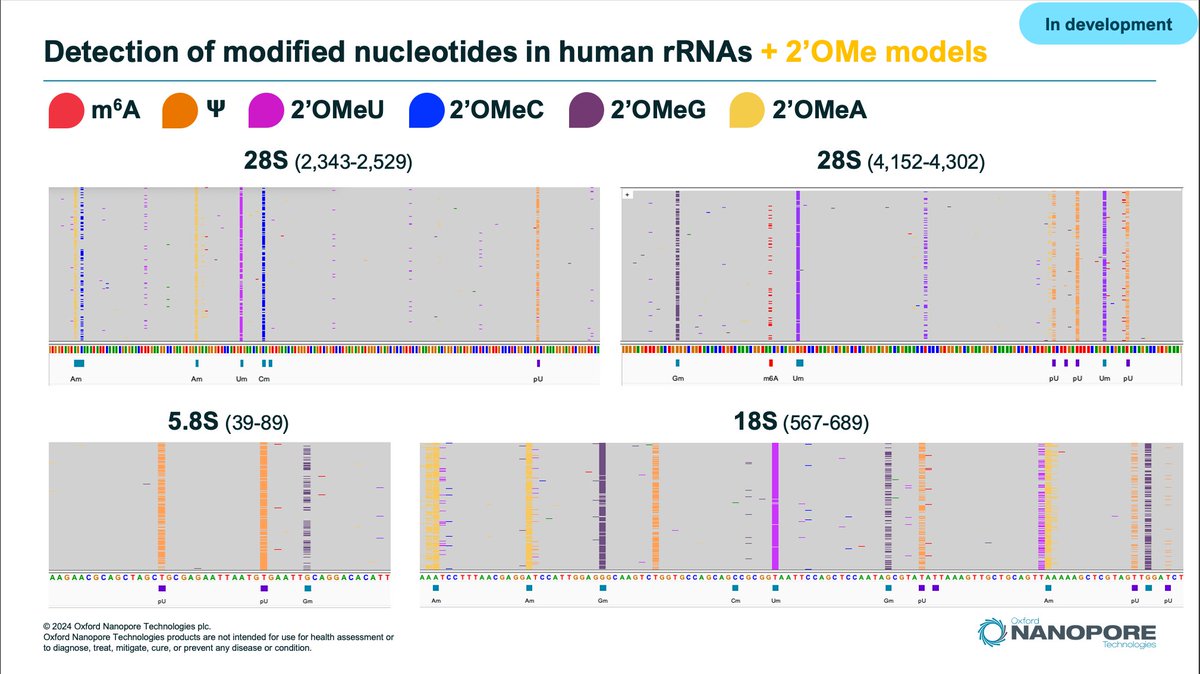
We have been busing working on research models to detect all 2'Ome-RNA modified nucleotides on top of PseudoU, m6A, m5C and Inosine using Oxford Nanopore dRNA seq. Preliminary results on Human #rRNA are very exciting. See🦋for more details bsky.app/profile/adrien…

Who has DNA methylation on ~5,000 microbes sequenced w/Oxford Nanopore? We do! Who’s working hard on getting this data out before #ASMicrobe in LA next month? We are!🙃 Stay tuned! Major updates coming ATCC Genome Portal starting w/methylation data on 1000’s of microbes! #Genomics



Tweet 1/2 WOW—2’O-methylation detection in direct RNA-seq is here! A breakthrough for RNA therapeutics, viral evasion, and mRNA stability. Oxford Nanopore now enables single-base resolution of RNA mods like 2’OMe-A/C/G/U directly from native RNA. #nanoporeconf Oxford Nanopore


ONT Oxford Nanopore is the only sequencing tech capable of direct RNA seq (without cDNA conversion step). Not only that, ONT can detect RNA modifications in the nucleotide bases and sugar! #Nanoporeconf
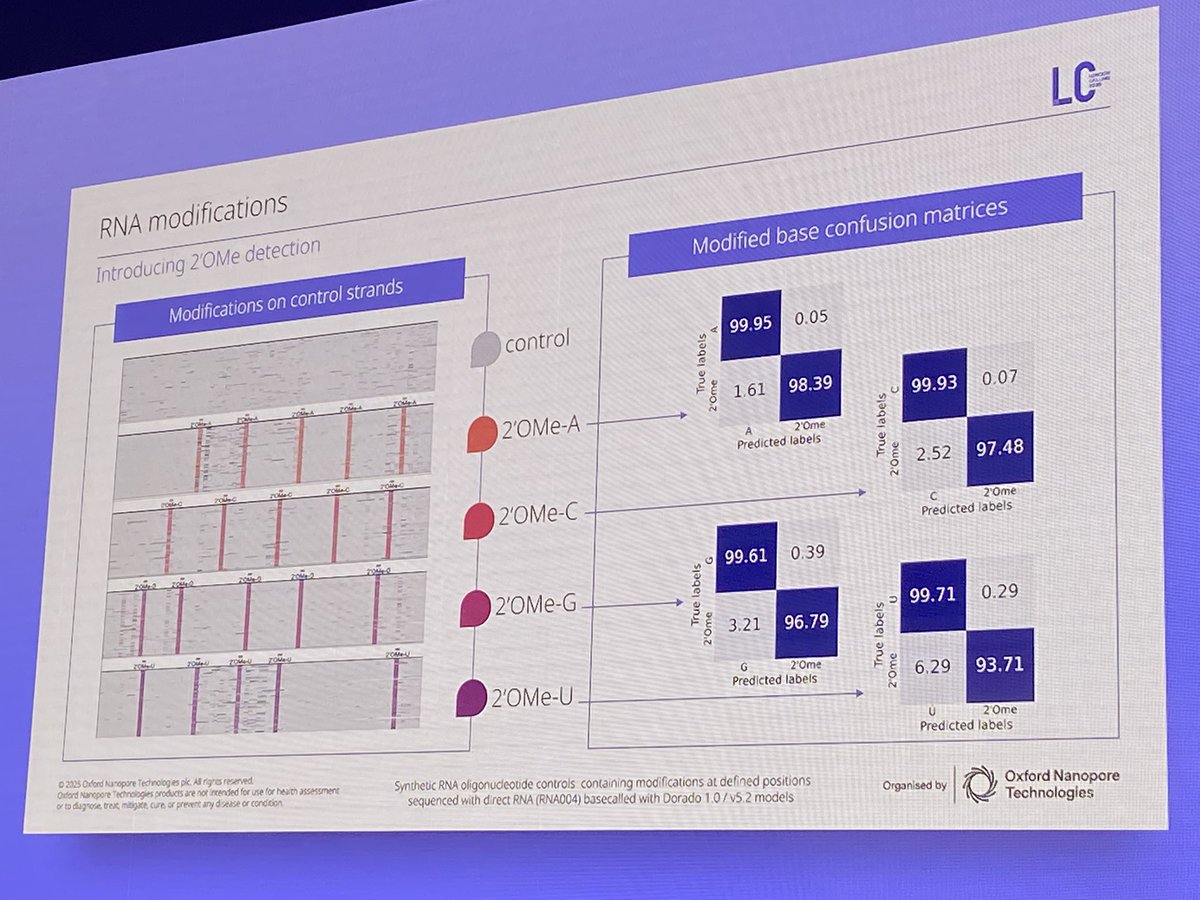

More details on new RNA modified bases detectable with Oxford Nanopore sequencing. #NanoporeConf





