
yinxiu zhan
@yinxiuzhan
Data data data
ID: 889403734232576000
24-07-2017 08:35:49
27 Tweet
64 Followers
61 Following

First deep learning project. Great teamwork with Bastian Eichenberger

Finally out in a peer-reviewed journal Genome Biology! PhD project of our first student Oriana Genolet - great work, we are really missing you in the lab! Great collaboration with Michael Boettcher and important contributions from Anna Alessandra Monaco! genomebiology.biomedcentral.com/articles/10.11…

Come join the fantastic Kim Jensen Lab in Copenhagen! An amazing group of scientists and a great supervisor!




Debutta a Milano la scuola per gli imprenditori deep tech startupbusiness.it/debutta-a-mila… Startupbusiness

1/5 I'm very proud to share our new story about the reconstruction quality of 3D chromosome conformation. Thanks to wonderful collaboration: Aleksandra Galitsyna, Sergey Ulianov, Mikhail Gelfand, Sergey Razin, and Alexander Chertovich (mspslab.com). doi.org/10.1371/journa…

What happens to gene regulation when an almost entire TAD is inverted? Check out our work below 👇 Heard_Lab @LucaGiorgetti yinxiu zhan Elphege Nora Lab at UCSF @Joke_van_Bemmel @maudborensztein a great team effort as usual :) thank you all 🙏

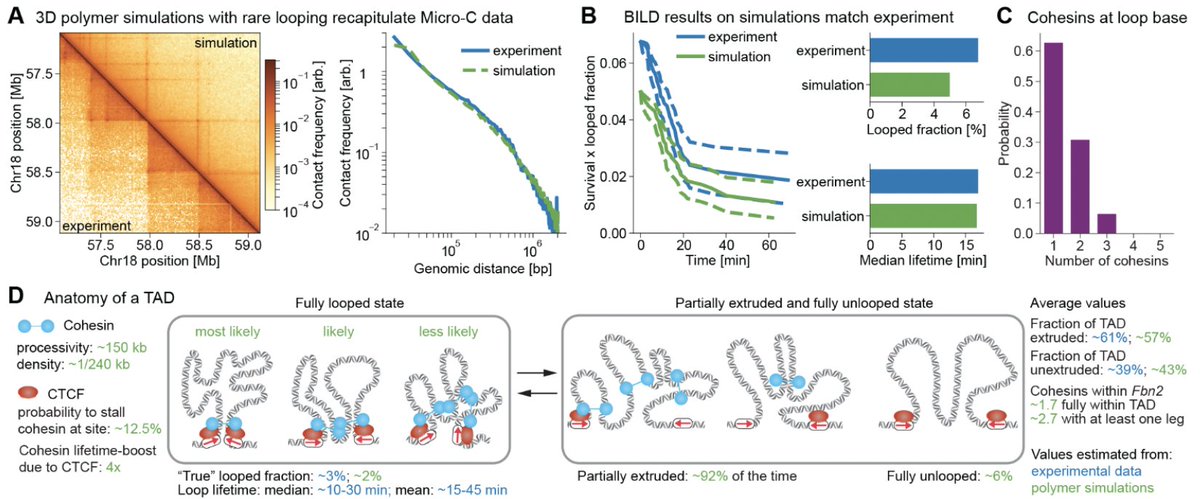
(1/6) our preprint biorxiv.org/content/10.110… is now out in Science Magazine science.org/doi/10.1126/sc… Using super-res live-imaging we show that CTCF/cohesin loops are dynamic (~10-30 min median lifetime), rare (~3-6.5% looped fraction), and we fully parameterize a TAD.



Congrats and well deserved! I have been very lucky to have had you as mentor (within and outside science!) and friend! Good job FMI science


A short Tweetorial on LAMMPS. LAMMPS (Sandia National Labs) is a v powerful engine to perform molecular dynamics simulations. Albeit popular, there are still some people who struggle to pick it up due to a relatively steep learning curve. I hope this will help you getting started.. (1/n)


In this episode we talked to @LucaGiorgetti from the FMI science to hear about his work on long-range transcriptional control by 3D chromosome structure. #chromatin #podcast #epigenetics Listen here: activemotif.com/podcasts#luca-…


Finally out, great collaboration with Lab Nicola Iovino. Using imaging, cut&tag and modelling, we showed that H3K9me3 is dispensable for HP1a recuitment during early development in drosophila!



