Walter Muskovic
@waltermuskovic
Post-doctoral researcher @ Garvan Institute of Medical Research, using single-cell sequencing to investigate the brain cancer glioblastoma.
ID: 3282194546
http://waltermuskovic.com 17-07-2015 07:24:37
168 Tweet
143 Takipçi
163 Takip Edilen



📢Oral abstracts for #ozsinglecell Perth Closing soon! Attention Tumor-immunology tweeps, talks from 👇 Miriam Merad, MD, PhD @realFlorentGinhoux @dralexswarbrick eventbrite.com.au/e/oz-single-ce…


Excited to share our recent work using 'villages' for large-scale stem cell studies. Only possible thanks to effort from Angela Steinmann, Han Chiu, Maciej Daniszewski and many more from Nathan Palpant, @AlicePebay, Joseph Powell labs and G-W Centre. biorxiv.org/content/10.110…

We are recruiting group leaders! Come and join me and set up your own research group at Peter Mac Research

Interested in lncRNAs? Check out our new paper up on bioRxiv where Prof Maria Kavallaris AM 🌈 and I use high temporal resolution RNA-seq time series data to demonstrate both human and mouse lncRNAs mirror the expression of nearby protein-coding genes doi.org/10.1101/2021.0…

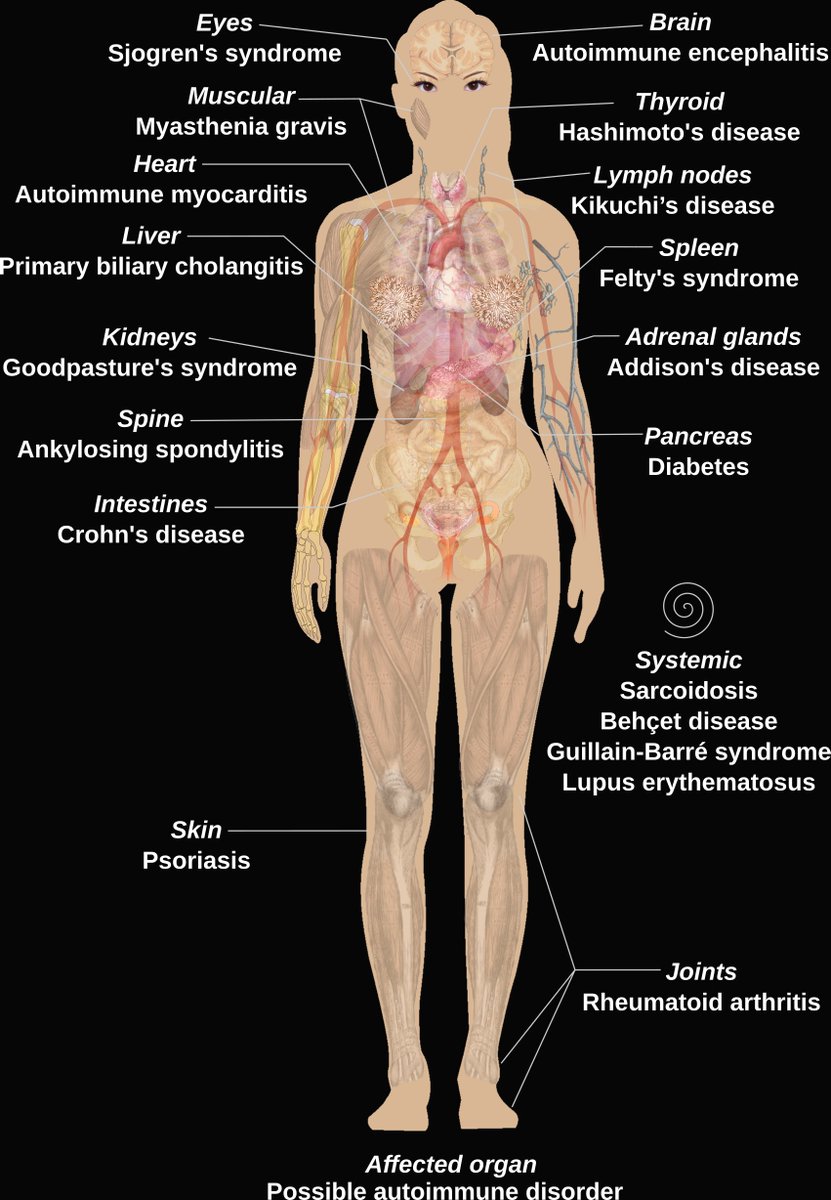
Living with an #AutoimmuneDisease or know someone who is? Check out Anna Liza Kretzschmar'stwitter presentation today at 2:45 as part of #SVCRS2021 and check out the autoimmune community survey spoonie-community.netlify.app #Spoonie #ChronicIllness #Autoimmune



Introducing Demuxafy: Improvement in droplet assignment by integrating multiple single-cell demultiplexing and doublet detection methods. Work led by Drew Neavin as part of the sceQTL-gen consortium biorxiv.org/content/10.110…





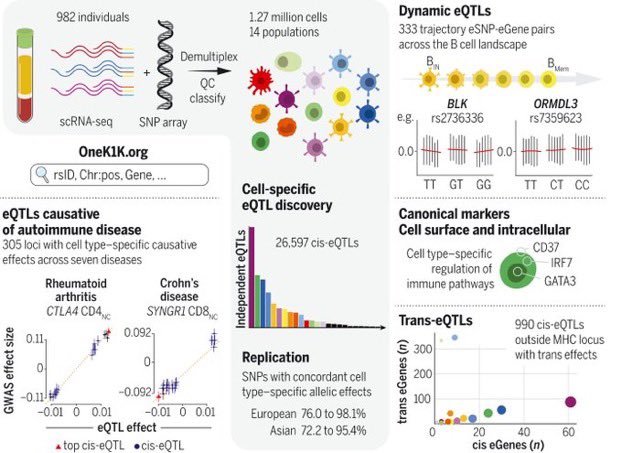
Our work “Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease” just published in Science Magazine. The work started with the creation of the OneK1K cohort, consisting of scRNA-seq data on PBMCs from ~1000 donors science.org/doi/10.1126/sc… 🧵

Excited to share our work on single cell eQTL mapping of OneK1K Cohort (1000 individuals 1000 cells) that’s just published in Science Magazine. Thanks to our amazing team José Alquicira Hernández, @WingKristof, Kirsten Fairfax, Gracie Gordon, Jimmie Ye, Alex Hewitt and Joseph Powell!

Many intercellular communication systems (BMP, Notch, Wnt, FGF, etc) use sets of promiscuously interacting ligand and receptor variants rather than seemingly simpler one-to-one architectures. Why? Our 2 papers grappling w/this question just out in Cell Systems



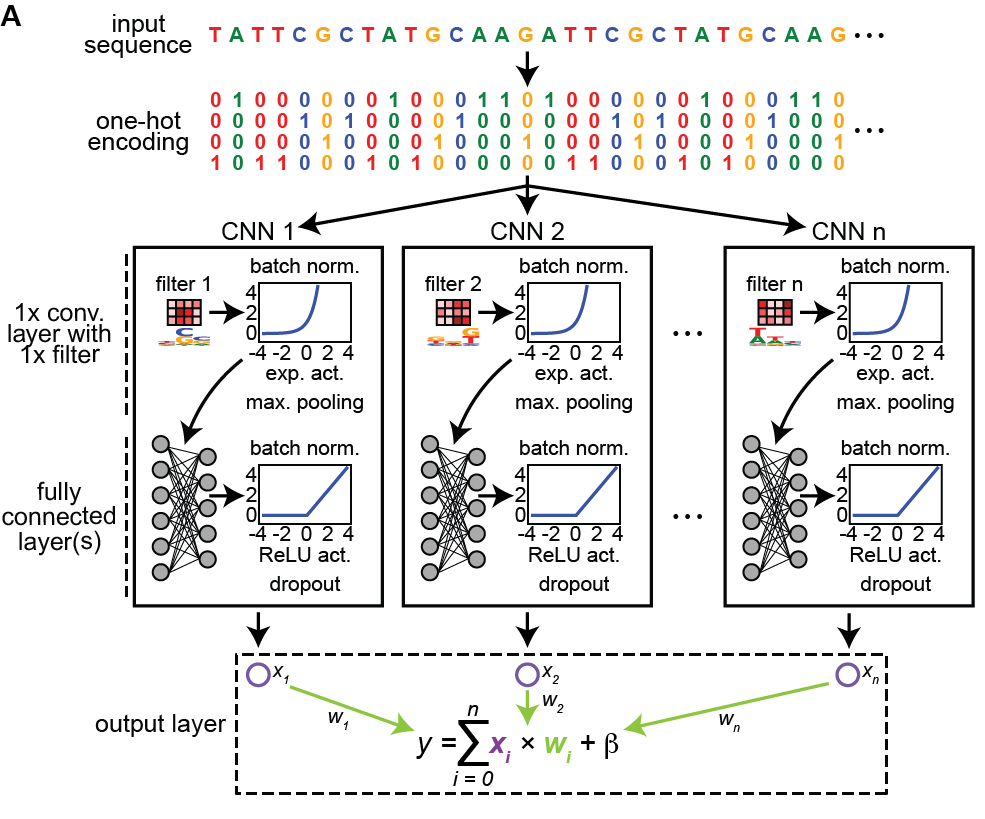
Want to apply #deeplearning in your #regulatory #genomics analyses but overwhelmed by suite of interpretation tools? Check out ExplaiNN: interpretable and transparent neural networks for genomics with Gherman Novakovsky (слава Україні! 🇺🇦) Oriol Fornés Sara Mostafavi Wyeth Wasserman 🧵👇 biorxiv.org/cgi/content/sh…