
Sven Eyckerman
@vibeyckermans
Expert Scientist @VIB, Associate Professor @UGent, Biochemistry, Signal Transduction, Protein Complexes, Proteomics, Cancer Pathways, Molecular Tinkering
ID: 135456352
21-04-2010 10:40:17
1,1K Tweet
389 Takipçi
389 Takip Edilen

.Broad Institute CeVICA, a cell-free nanobody engineering platform that uses ribosome display and computational clustering analysis for in vitro selection. go.nature.com/3AlD44n

So excited with the approval of our UGhent GOA REINFORCE program aiming to study the interface of innovative drugging strategies and tumor microenvironment in neuroblastoma. Congrats to all the amazing team members Jo Vandesompele Charlotte Scott @Svennetti Steven Goossens Katleen De Preter

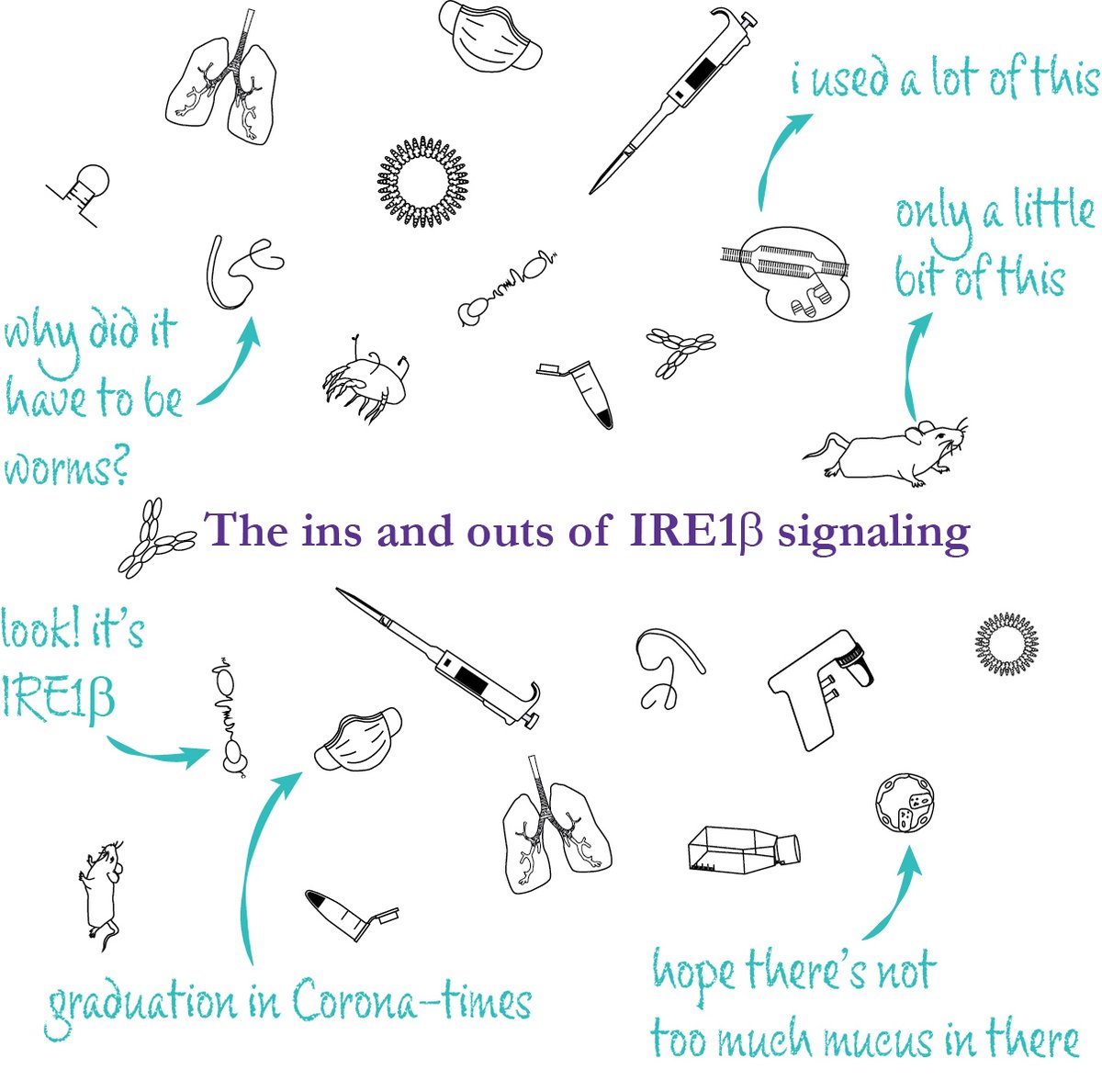
About seven years ago I came eye to eye with a little protein called IRE1β when I joined the @Svennetti and Sophie Janssens Lab labs. Welcome to The Ins and Outs of IRE1β signaling: the Thesis Thread (1/16) #ThesisThread #ScienceCommunication VIB Ghent University Research


Does your research rely on recombinant protein production? Check out this new method to predict protein secretability by @NicoCallewaert, Morgane Boone, CompOmics, Wesley De Neve & colleagues VIB, Ghent University Research & VUB. 👉🏼Today in Nature Communications vib.be/news/new-metho…










Two papers from OncoRNALab out in special issue on "Bioinformatics methods to study circular RNAs" doi.org/10.3389/fbinf.… and frontiersin.org/articles/10.33… Congrats Marieke Vromman and Annelien Morlion

📢 #JustPublished: Recent work from Saba Safdar, PhD, Seppe Driesen, @KarenLeirs and @Svennetti describing how the combination of nucleic acid engineering and CRISPR can be used for innovative #biosensing concepts.🧬 👉Read: doi.org/10.1016/j.bios… FWO Marie Skłodowska-Curie Actions KU Leuven









