
Willem van Rengs
@rengswillem
Researcher plant genetics and genomics | Experience in experimental and computational biology | PhD | MPIPZ alumni | Music lover
ID: 1395347663453818880
20-05-2021 11:56:31
153 Tweet
185 Followers
292 Following

Want to hear something on #banana #sequencing and #genomics? Don't mis out on Alexander Wittenberg talk on banana pangenome construction at the #nanoporeconf Oxford Nanopore London calling.

Happy to present banana 🍌 pangenome using Oxford Nanopore duplex data at #nanoporeconf The insights obtained help in breeding new varieties that are resistant to Fusarium Tropical Race 4 (TR4) and Black Sigatoka. Thanks Willem van Rengs for the assemblies and comparisons.



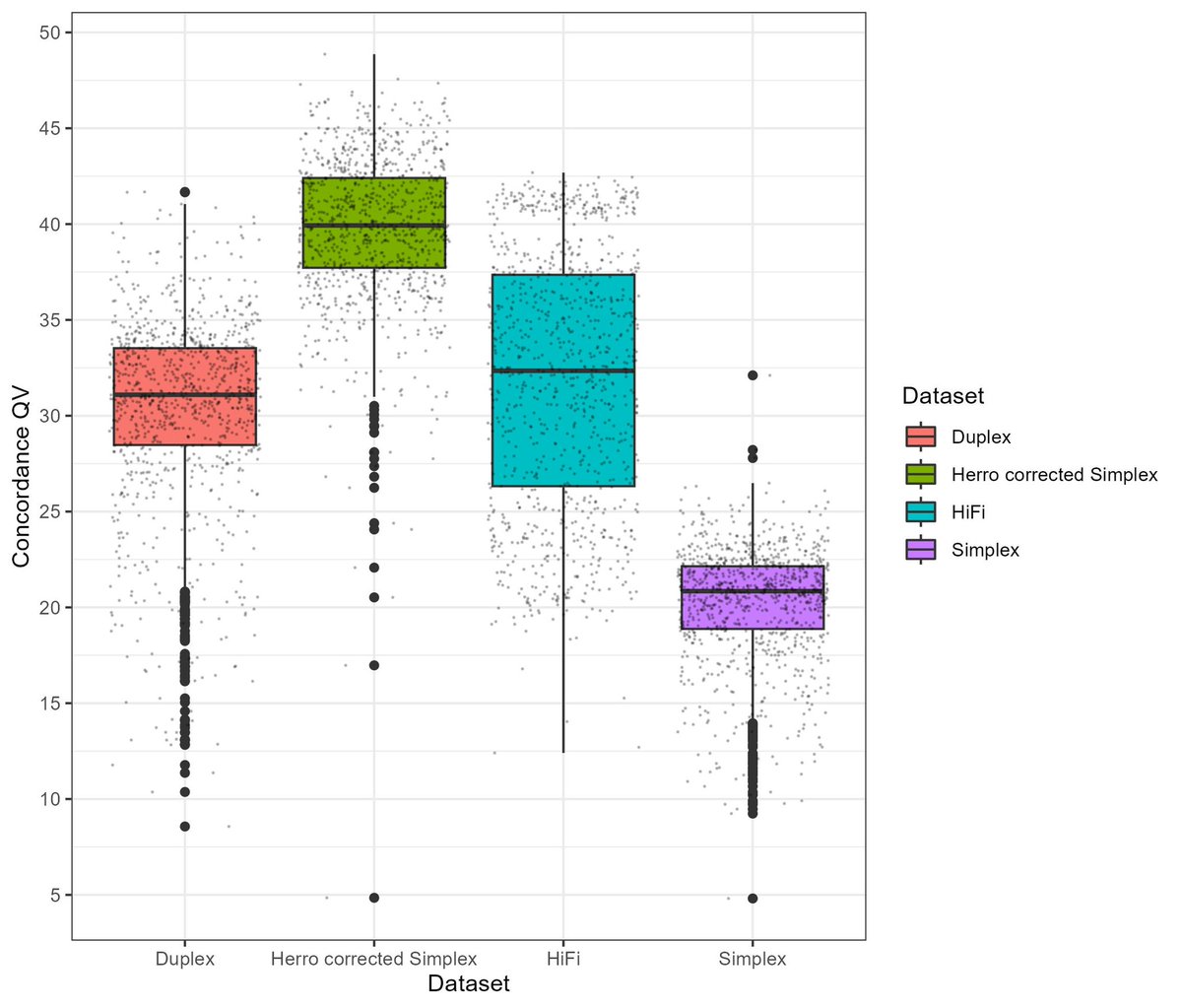
Herro error corrected @keygene banana 🍌 Oxford Nanopore simplex reads versus duplex and HiFi data. Amazing work by Mile Sikic and team. See you next week in London! #nanoporeconf #accuracy #pangenome #banana

Associated research briefing written by Yazhong Wang and I is also now published: rdcu.be/dIExV




Looking forward to present in the Oxford Nanopore workshop during PAG-International Plant & Animal Genome Conference Australia in three weeks; “ Unveiling the Banana 🍌 Pangenome: A Leap Towards Sustainable Cultivation and Enhanced Disease Resistance” #banana #Diversity #pangenome #TR4 #keygene pag.confex.com/pag/au2024/mee…



In just one week I will be representing @keygene presenting at #PAGAuatralia int he Oxford Nanopore workshop on the banana 🍌 pangenome; pag.confex.com/pag/au2024/mee… Looking forward to catch up!



The latest hifiasm can directly assemble standard Oxford Nanopore simplex R10 reads, without HERRO correction or other preprocessing, to phased contigs of contiguity comparable to HiFi assembly. Like before, you can further add ultra-long, Hi-C or trio data for better assembly.

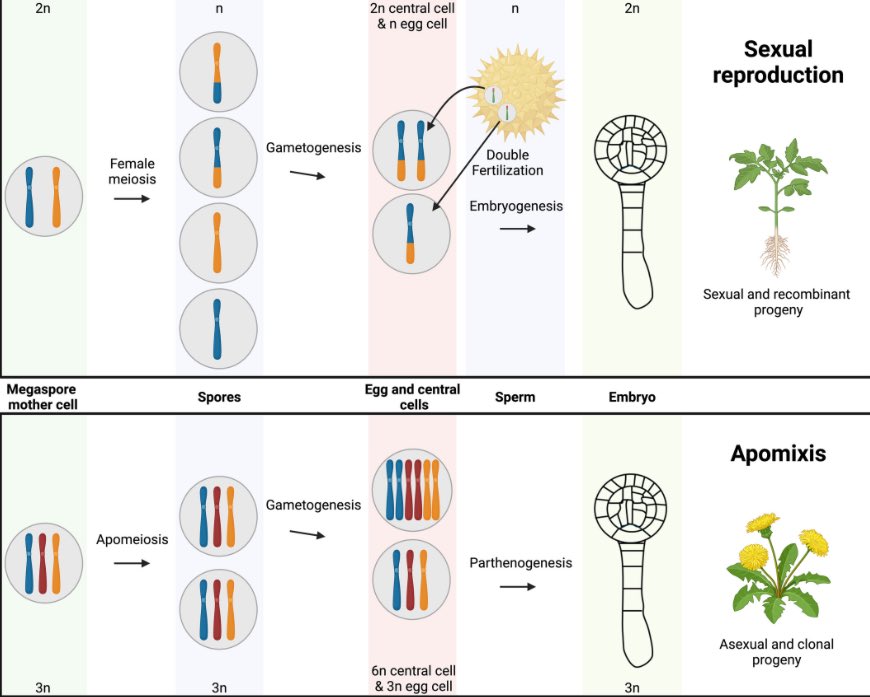
Our new review article “To infinity and beyond: recent progress, bottlenecks, and potential of clonal seeds by apomixis” is now published open access in The Plant Journal dx.doi.org/10.1111/tpj.... Radboud University






Tomorrow Luciana Gaccione from Lorenzo Barchi' lab will explain on our Oxford Nanopore day at HHU Düsseldorf, how to turn 9.41 data(!) into a high quality pangenome. If you can't make it read all about it here as well: nature.com/articles/s4146…
