Rafael Josip Penić
@rjpenic
PhD student @ Faculty of Electrical Engineering and Computing, University of Zagreb | 🇭🇷 | Machine Learning | AI in Structural Biology
ID: 1484504640414699526
21-01-2022 12:34:34
31 Tweet
43 Followers
149 Following


Long reads metagenome benchmark is out. Highlights. 1. In most cases Kraken, 2. minimap2/ram for slightly higher accuracy. 3. The right database is of huge importance 4. Check taxonomy files carefully. bmcbioinformatics.biomedcentral.com/articles/10.11… w/Niranjan Nagarajan Krešimir Križanović @jmaricb Sylvain

Are nanopore UL reads only long reads we need? We developed Herro github.com/lbcb-sci/herro AI error correction model that can correct reads to accuracy above Q30 while trying to keep informative positions intact. w/ Dominik Stanojevic Dehui Lin Sergey Nurk 🇺🇦 Pore_XX_Singapore Oxford Nanopore





Rockfish: A transformer-based model for accurate 5-methylcytosine prediction from nanopore sequencing has been published in Nat. Comm!! Great work by Dominik Stanojevic w/ Zhe Li, Sara Bakić and Roger Foo Paper: nature.com/articles/s4146… Code: github.com/lbcb-sci/rockf…

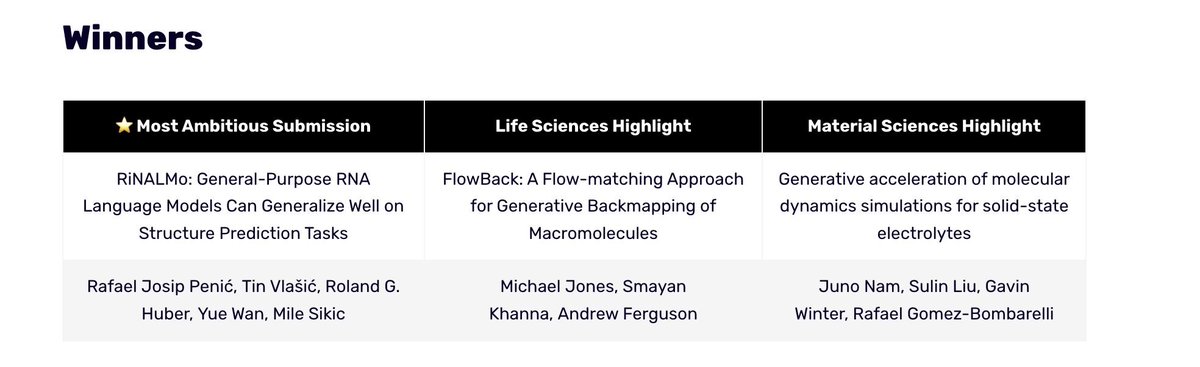
Our RNA LLM RiNALMo has received the prize for the most ambitious submission at the Machine Learning for Life and Material Science workshop at ICML Conference 2024. w/ Rafael Josip Penić Tin Vlasic Yue Wan and Roland Huber. Workshop: ml4lms.bio Preprint: arxiv.org/abs/2403.00043




RNA structure prediction is still an open problem. Look at our benchmark results (including Alphafold 3!) biorxiv.org/content/10.110… w/ Ivona Martinović Tin Vlasic Yang Li Bryan Hooi and Zhang Yang





I am happy to share our new preprint introducing MADRe - a pipeline for Metagenomic Assembly-Driven Database Reduction, enabling accurate and computationally efficient strain-level metagenomic classification. Mile Sikic, Riccardo Vicedomini, Krešimir Križanović 🔗biorxiv.org/content/10.110… 1/9

I am happy to introduce Campolina, a deep neural framework that replaces traditional algorithmic approaches for nanopore signal segmentation and improves segmentation quality for real-time analysis. Preprint and details in the thread👇 K. Friganovic, Bryan Hooi, Mile Sikic 1/7