James Ferguson
@psy_fer_
Bioinformatician/Genomics Software Engineer @GarvanInstitute, Views my own.
Mastodon @[email protected], genomic.social
BS: psy-fer.bsky.social
ID: 761420960046080000
http://jmferguson.science 05-08-2016 04:37:58
14,14K Tweet
3,3K Followers
1,1K Following




Long reads Oxford Nanopore PacBio can provide insights into epigenetics. We (Yilei Fu Winston Timp) summarize the latest #Bioinformatics approaches to analyze methylation & why this matters! Out now Nature Reviews Genetics bit.ly/4cjxUtl BCM HGSC BCM Department of Molecular and Human Genetics Rice Computer Science Johns Hopkins University


Introducing cornetto, an adaptive genome assembly paradigm using Oxford Nanopore adaptive sampling. - greatly reduces cost per genome assembly - reference agnostic, so works for non-humans - assembly just using saliva - & many more Relies on 2 excellent software #readfish & #hifiasm.

A/Prof Jodie Ingles and A/Prof Owen Siggs spoke to 7NEWS Sydney recently about the launch of the Rare Disease Registry at Garvan. This program aims to improve diagnosis and treatment options for the two million Australians living with a rare disease. ow.ly/xtkx50VvkXH




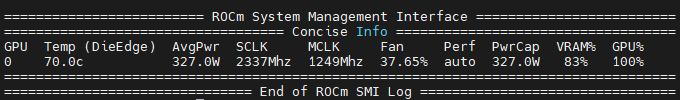
Realfreq, a framework for real-time base modification analysis for nanopore sequencing, is now published in Bioinformatics. Written efficiently using C, a laptop can keep up with a Oxford Nanopore MinION sequencer, and a desktop a PromethION 2 solo flowcell. academic.oup.com/bioinformatics…









A new compression strategy to reduce the size of nanopore sequencing data. #DataCompression Oxford Nanopore #SequenceData #Bioinformatics #Genomics Genome Research genome.cshlp.org/content/early/…


Recently, I sat down with Sebastian Cocioba (Sebastian S. Cocioba🪄🌷) to chat about what real-time science can look like in practice. He spoke about his open lab-notebook, Flowers for Everyone, the necessity of documenting failures, and what it takes to run a one-man CRO today. (1/2)