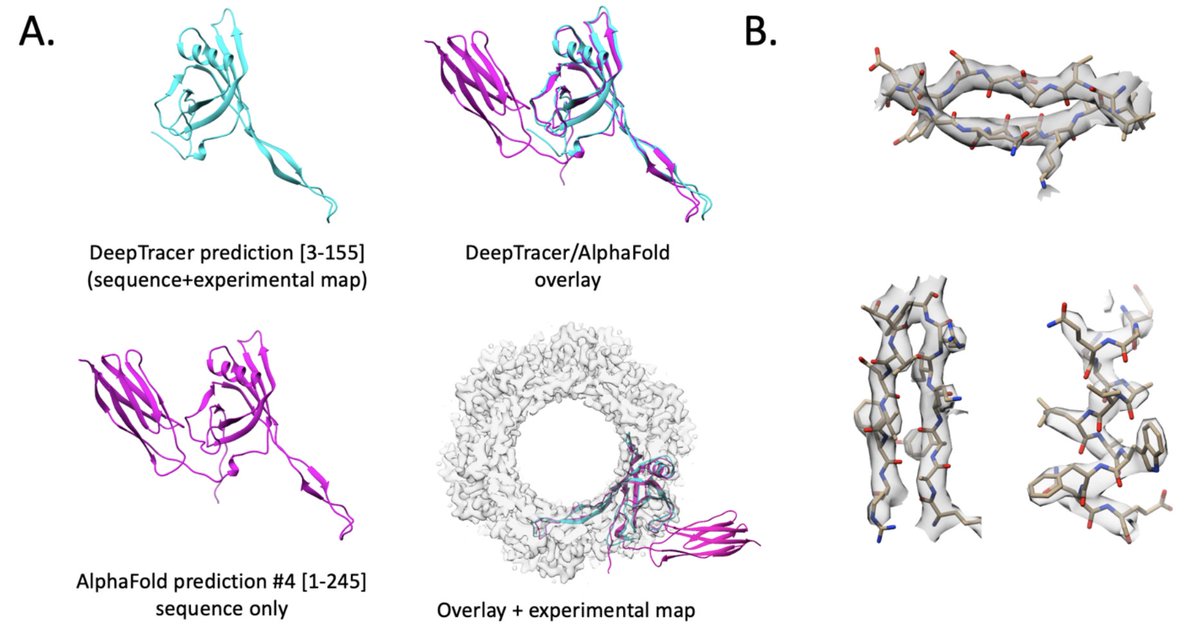
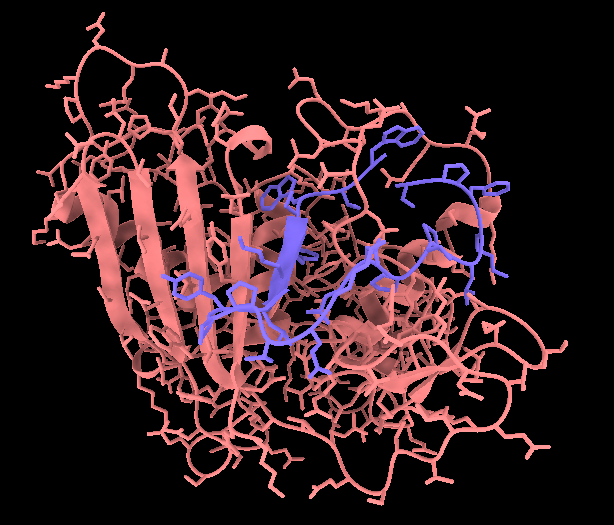
Niklas Rindtorff
@niklas_tr
assembling molecular rings @ convexity labs. past @dkfz | @BroadInstitute | @HarvardDBMI
ID: 3753717148
http://rindtorff.xyz 23-09-2015 22:17:02
1,1K Tweet
3,3K Followers
3,3K Following






New #AI-ready dataset = a leap forward for drug discovery & energy technologies. AI at Meta + Berkeley Lab co-led #OMol25: the most diverse open-source chemistry dataset ever built. Science just got a major upgrade. @lbnlcs Berkeley Lab ETA #OpenScience newscenter.lbl.gov/2025/05/14/com…






Seems like a lot of the pre- vs. post-training compute requirements can be boiled down to "The lower the rate of observing useful training information, the lower the bandwith between nodes can be." Also great to see Prime Intellect getting a shoutout!