Kirubakaran Palani
@npkirubakaran
Institute Research Investigator | MD Anderson Cancer Center | Computational Chemistry and Drug Design | Deep Learning 🖥💻💊🔬💉
ID: 1110762091
https://www.mdanderson.org/research.html 22-01-2013 06:19:06
69 Tweet
74 Followers
308 Following

Very proud of this work led by Aveda.Sci Elias Ndaru, Ph.D. and Alisa Garaeva with fantastic collaborators Cristina Paulino Max Bonomi, Dirk Slotboom, and Christof Grewer. We use modeling, cryo-EM, synthetic chemistry, and functional assays to characterize ASCT2 biorxiv.org/cgi/content/sh…



How is a metal formed? An int. team led by Pavel Jungwirth #IOCBPrague has mapped at the molecular level the electrolyte-to-metal transition in alkali metal–liquid ammonia solutions and earned cover page in Science Magazine. uochb.cz/en/news/182/ho… doi.org/10.1126/scienc…





Learning how to sell a story to ANY audience is an important skill to learn. Check out this latest #scicomm effort by Fox Chase Cancer Center postdocs. Kudos to Bailee Sliker, Ph.D, Kirubakaran Palani, AlejandraV Contreras, Alyssa Leystra, Uttam Satyal, Cristina Uribe, and Rob Sertori!

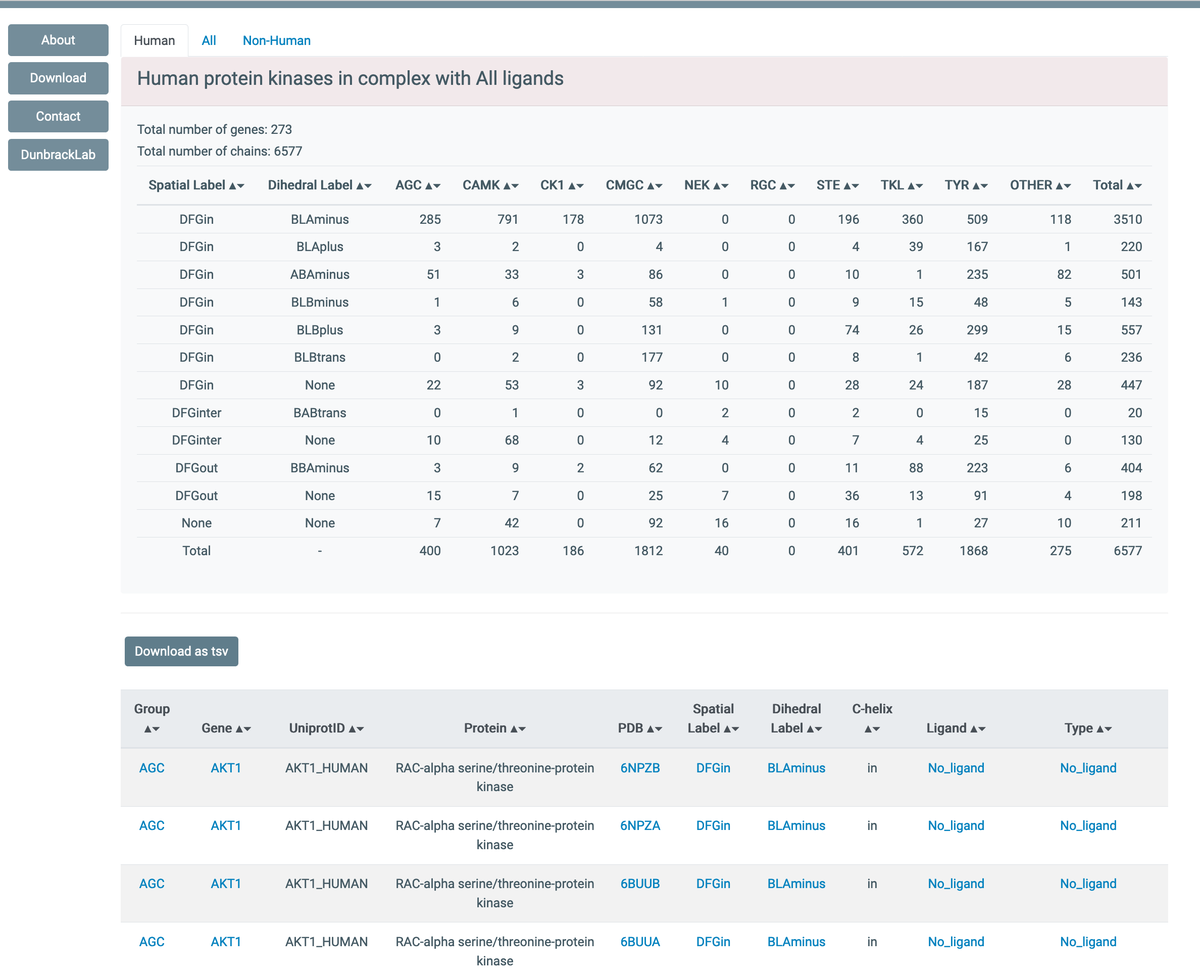
Our database on the classification of protein kinase conformations is now online:dunbrack3.fccc.edu/kincore Browsable by kinase gene/family, conformational class (6 DFGin classes, 1 DFGout, 1 DFGinter), PDB ligand, inh. type. Appreciate feedback/RT. rcsb pdb 💉🧬💻🔬💊🌱🧠🦠 Protein Data Bank PDBj_en



Join us tomorrow at @ 4 PM for an online seminar featuring Roland Dunbrack Roland Dunbrack 🏳️🌈 @rolanddunbrack.bsky.social of Fox Chase Cancer Center, hosted by James Fraser (A$AP J)! Register here and tune in: qbi.ucsf.edu/seminar-dunbra…



We created a website & Python script that renumbers residues in PDB files with UniProt numbering. It returns mmCIF & PDB format files of the ASU & bio assemblies for any list of entries or UniProtIDs. Plz help us test it. rcsb pdb 💉🧬💻🔬💊🌱🧠🦠 PDBj_en Protein Data Bank dunbrack3.fccc.edu/PDBrenum/



Excited to make this important work public! It turns out that building *truly useful* AI models for inhibitor/kinase interactions remains really challenging, as demonstrated by the careful studies of Gabriel Ong and Kirubakaran Palani. biorxiv.org/content/10.110…