
Mike Liu
@mike_c_liu
Head of Operations and Product Development Lead at Molecular Instruments @HCRimaging
ID: 1011965408205627392
27-06-2018 13:32:09
126 Tweet
412 Takipçi
836 Takip Edilen

This week’s #FluorescenceFriday user is Dominic Shayler! #HCRimaging enabled Dominic to validate his single-cell RNA sequencing data through reliable ISH results, even from poorly characterized genes. Check out his remarkable HCR™ RNA-FISH images of human retinal tissue here:


mapzebrain, the open-source atlas of the zebrafish brain, now also contains detailed gene expression maps. The atlas allows seamless integration of diverse data modalities, making it an important research tool accessible to everyone. science.org/doi/10.1126/sc… Inbal Shainer 🎗️ @Enmyco


Thank you Lisanne Schulze, David Zada and Matt Lovett-Barron for highlighting our work. Proud to see our paper and resource out and hope it will help advancing zebrafish research in the years to come. Go Fish! 🐟 science.org/doi/10.1126/sc…

This week’s #FluorescenceFriday user is Isabel Holguera! #HCRimaging of deadpan mRNA (blue) allowed Isabel to identify neural stem cells in the Drosophila optic lobe (outlined with a white dashed line) and central brain. Hoechst (orange) marks cellular doi.org/jxrz


This week’s #FluorescenceFriday users are Madeline Ryan and Tony Yu-Chen Tsai! Through #HCRimaging, Madeline and Tony were able to map out cell-type specific expression of various adhesion molecules in the zebrafish spinal cord. Check out their stunning images #withHCR here:




I am so happy to share the main portion of my PhD work AmphiSpace, out now in preprint! 🎉 We investigated the evolution of the anterior neuroectoderm to trace the origin of the #forebrain. What did we find? Have a look at this tweetorial to find out! 🧵 tinyurl.com/53pbph8t

The making of chelicerate appendages: Dual Functions of labial Resolve the Hox Logic of Chelicerate Head Segments UW-Madison iBio Molecular Biology and Evolution #Hox #evodevo #arachnids academic.oup.com/mbe/article/40…


This week’s #FluorescenceFriday user is on Petra Popovics, a former lab member in @labRicke at University of Wisconsin O'Brien Center! #HCRimaging helped Petra Popovics identify different immune cell types in the mouse prostate and localize Spp1 expression in luminal microphages. doi.org/j265


This week’s #FluorescenceFriday user is Eric Horstick! #HCRimaging enabled Eric Horstick to test for gene expression and neural activity changes in vivo. View his beautiful HCR RNA-FISH images of larval zebrafish below, and check out his lab horsticklab.org




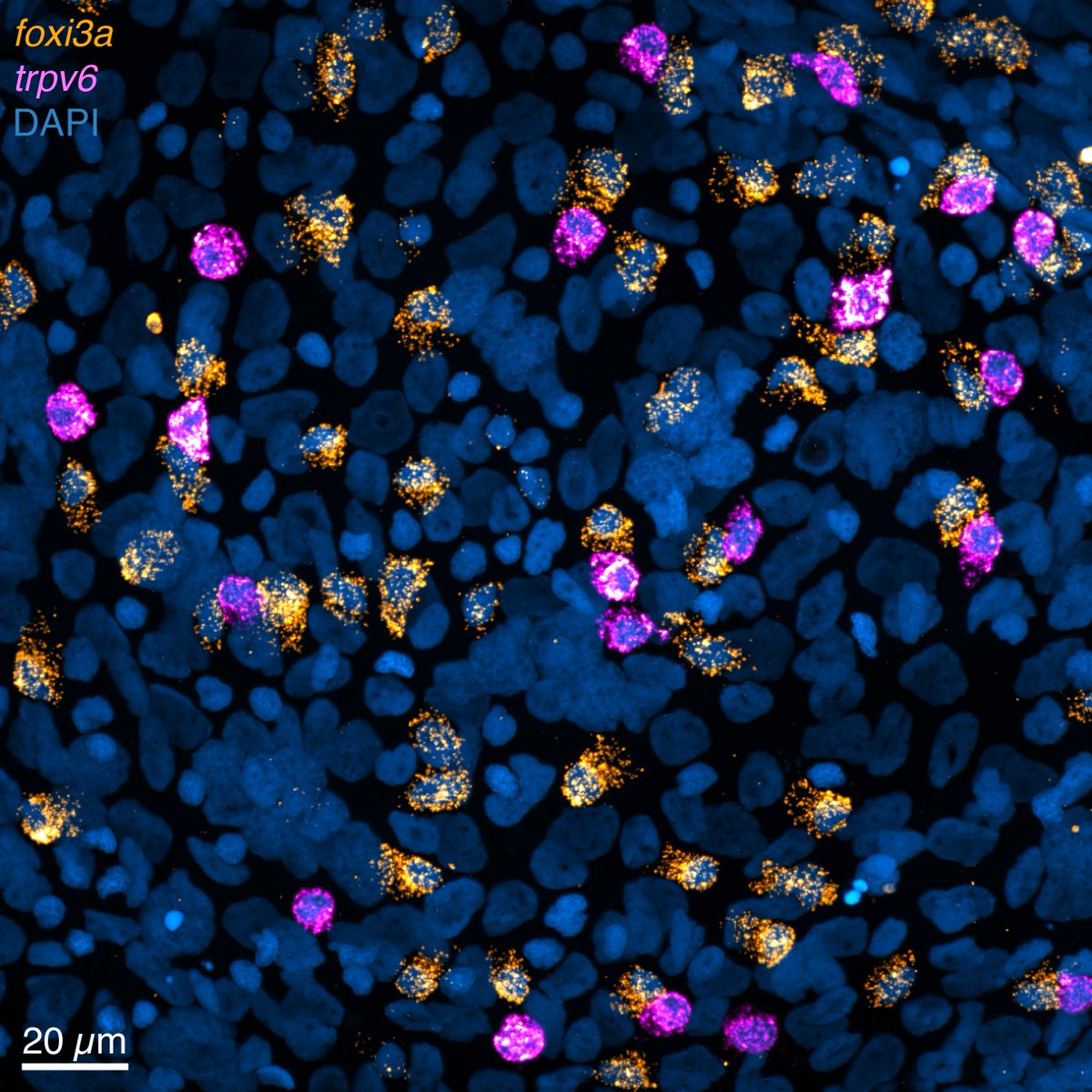
This week’s #FluorescenceFriday user is Julia Peloggia de Castro, a PhD candidate in Piotrowski Lab at Stowers Institute! @jupeloggia studies developmental plasticity using the zebrafish lateral line and ionocytes. #HCRimaging enabled her to map gene co-expression with single cell

Excited to share that our cuttlefish brain atlas is now out in Current Biology! Check out our website Cuttlebase.org and play around with the brain of this crazy camouflaging cephalopod. It was an amazing team effort 🐙✨ authors.elsevier.com/sd/article/S09…

This week’s #FluorescenceFriday user is Madeline Ryan, a research technician in the Tony Yu-Chen Tsai lab! Madeline Ryan uses #HCRimaging as a robust way to map cell-type expression patterns of different adhesion molecules and signaling pathway players in the developing zebrafish



This week’s #onlywithHCR Series features the Bedois et al. (2024) publication, “Sea lamprey enlightens the origin of the coupling of retinoic acid signaling to vertebrate hindbrain segmentation.” With #HCRimaging, Alice M. H. Bedois and her colleagues were able to investigate how







