PurdueMBTP
@mbtppurdue
T32 Funded Molecular Biophysics Training Program at Purdue, providing specialized training for graduate students interested in studying life through physics.
ID: 1120124131331203073
https://molbiophys.science.purdue.edu 22-04-2019 00:36:18
268 Tweet
173 Takipçi
34 Takip Edilen



Distinguished Professor Richard Kuhn named 2023 winner of Morrill Award, Purdue’s highest honor purdue.edu/newsroom/purdu… Richard J. Kuhn


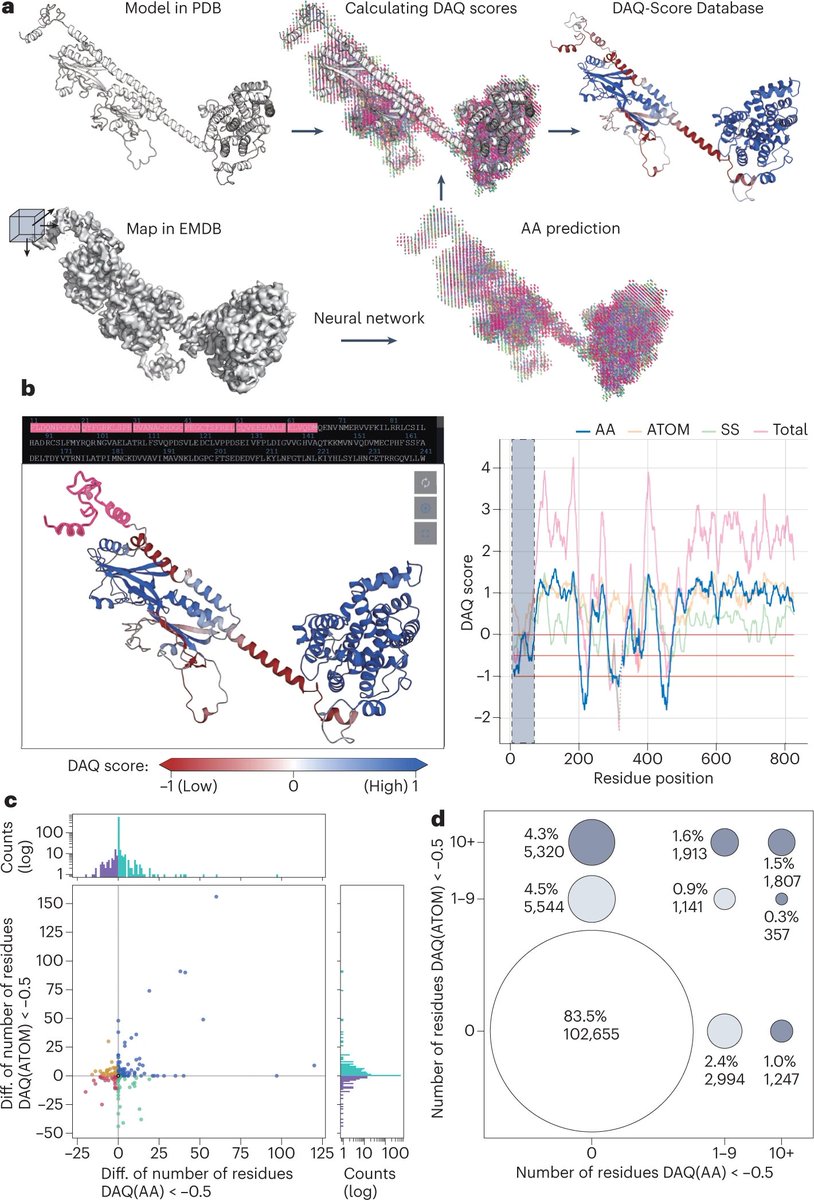
Just out online! "DAQ-Score Database: assessment of map–model compatibility for protein structure models from cryo-EM maps", Tsukasa Nakamura, Xiao Wang, Genki Terashi, & Kihara, Nature Methods Nature Methods nature.com/articles/s4159… Visit the db at daqdb.kiharalab.org

Congratulations, Stahelin Lab !!


Excited to have our grad students Shubham and Michelle present their outstanding research, collected with the help of NCCAT Info! Purdue Structural and Comp. Biology & Biophysics PurdueMBTP


We are looking for colleagues in chemical biology broadly defined at UMLifeSciences University of Michigan Reach out if you are interested in learning more about this awesome opportunity!! 🧪🧬🥼👩🔬

The The University of Chicago Department of Chemistry is currently accepting applications for several tenure-track Assistant Professor of Chemistry positions. All areas of chemistry are considered & joint appointments with other departments are possible. Apply now: chemistry.uchicago.edu/news/uchicago-…

New preprint online from the Stahelin lab and collaboration with the laboratory of Gregory Voth Voth Group lead by Mandira Dutta. Anionic lipid interactions of the SARS-CoV-2 nucleoprotein! biorxiv.org/content/10.110…

Our new paper accepted in MICCAI2023 is now online: "ACC-UNet: A Completely Convolutional UNet Model for the 2020s" Nabil Ibtehaz & Kihara from Purdue Computer Science link.springer.com/chapter/10.100…




Our Lyman Monroe Ph.D. appeared in an online career seminar! Thank you, Purdue Structural Biology and Biophysics Club, for organizing the seminar.



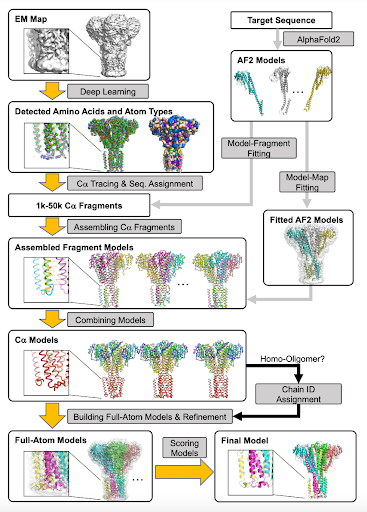
DeepMainmast is a protein structure modeling protocol for cryo-EM that combines the strengths of a deep-learning-based de novo protein main-chain-tracing approach with AlphaFold2-based structure predictions for improved performance. Genki Terashi doi.org/10.1038/s41592…