Lucy Colwell
@lucycolwell37
ID: 1124784990300950529
04-05-2019 21:16:53
20 Tweet
362 Takipçi
31 Takip Edilen



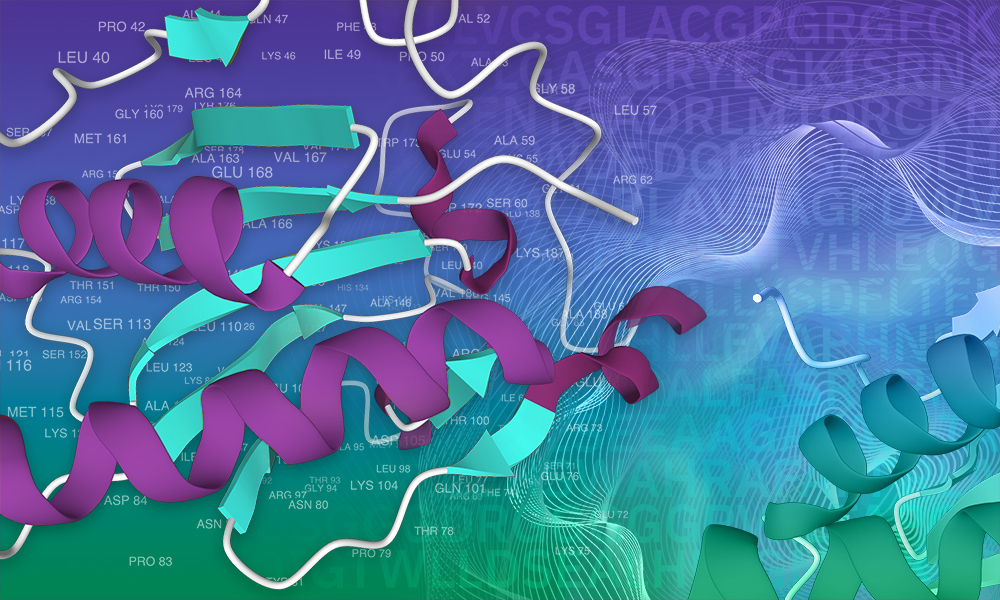
Predicting the function of proteins using #neuralnetworks can greatly aid medical research. We are proud to collaborate with EMBL-EBI to extend the The Pfam database database of functionally annotated proteins by 10% as part of the v34.0 release. Learn more below! xfam.wordpress.com/2021/03/24/goo…

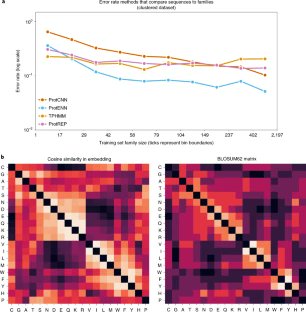
#AI is revolutionising protein science. Can it add to our knowledge of protein function? New Nature Biotechnology research shows how deep learning models can be used to improve protein annotations within The Pfam database and help predict protein function. nature.com/articles/s4158…



Nature Biotechnology Google AI The Francis Crick Institute MIT CSAIL EMBL-EBI Cambridge Chemistry The Pfam database Alex Bateman Lucy Colwell Thanks for all the interest everyone, and to the journal for the very helpful review. Here's a free link to the complete paper: rdcu.be/cHsqz


A smooth career path for many academics. nature.com/articles/s4156… 🙏Susanne Täuber Morteza Mahmoudi, PhD #bullying #whistleblowing #corruption


Machine learning can help read the language of life: blog.google/technology/ai/… Discussing our work in Nature Biotechnology, our interactive preprint, and why Google is involved.
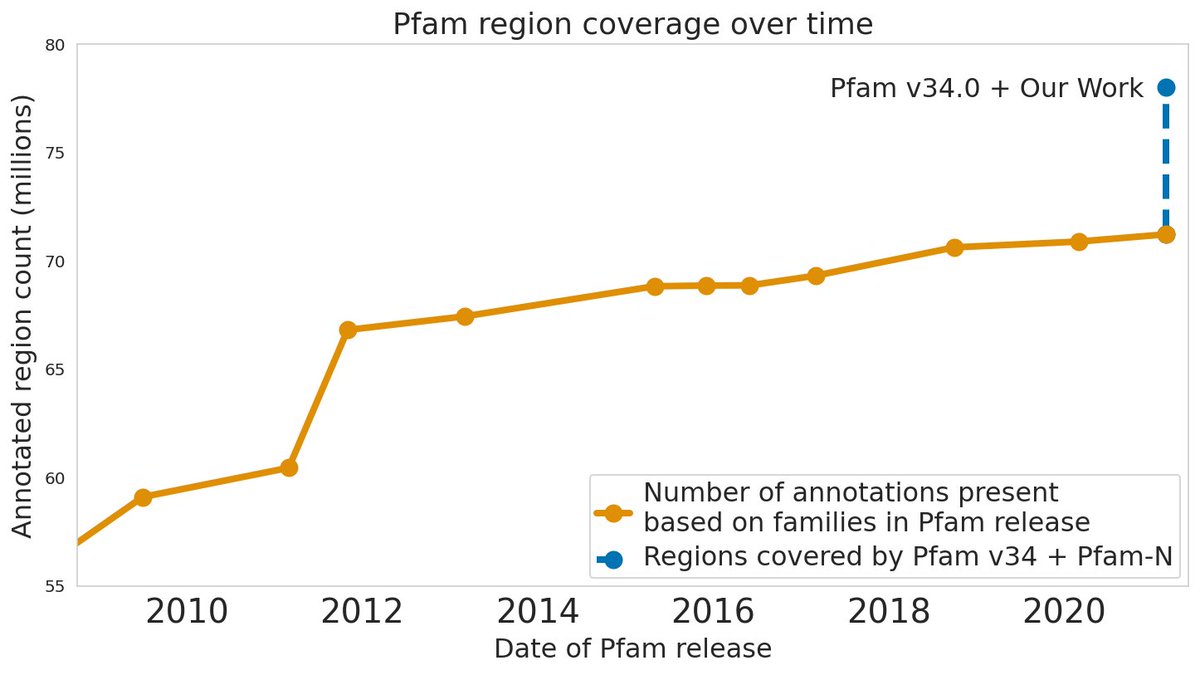



With help from #deeplearning models, The Pfam database has expanded by almost 10% exceeding all expansion made to the database over the last decade 🤯 Alex Bateman discusses this work and what the future holds for protein family classification. ebi.ac.uk/about/news/per…