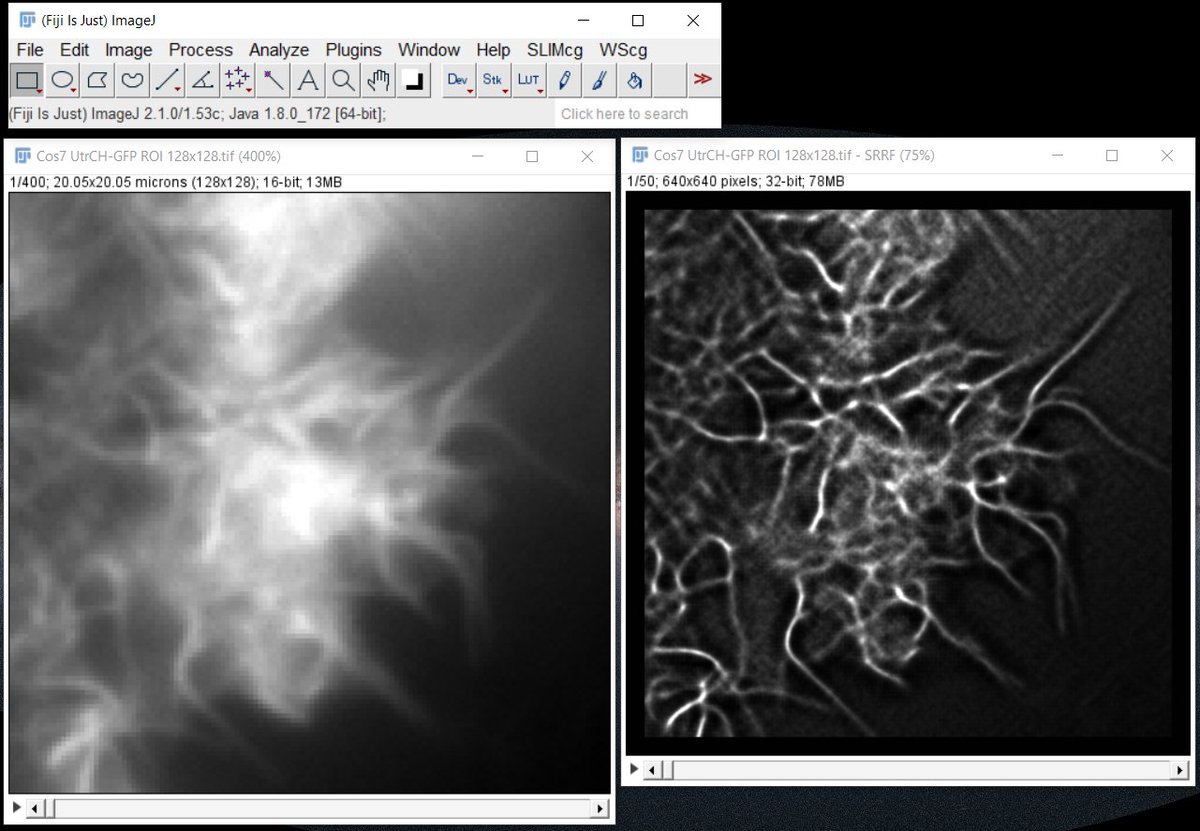
It is time to eSRRF 🏄♂️ at #FOM2024 with Hannah Heil Romain F. Laine Ricardo Henriques and also some advertisement for Single Molecule Localization Microscopy Symposium in Lisbon !



@NEUBIAS_COST - new webinar to happen on June 15th at 15h30-17h00 CEST (Brussels Time).
by Guillaume Jacquemet (ABO Academy, Turku) and Romain F. Laine (UCL, London)
#Webinar #NEUBIAS_Academy #DeepLearning #Microscopy


Happy to organise a single-day webinar w/GDR ImaBio ' #Imaging_viruses ' March 16th Register at : bit.ly/3jvEAJG speakers Eggeling Lab, Ricardo Henriques ,@SergiPadilla3 MURIAUX Delphine Romain F. Laine Christian Sieben Saveez Saffarian Nicole Robb.


I'm really excited that #ZeroCostDL4Mic is out and that I could contribute to this amazing mission of widening access to #DeepLearning !
Thanks @LaineBioimaging Lucas von Chamier Ricardo Henriques Guillaume Jacquemet for bringing me along!
👇Whole-cell mito segmentation with our 3D U-Net


The latest version of the Microscopy Icons are now on GitHub and available under CC-BY! Share and enjoy 😀
github.com/lacan/microsco…
Thanks Romain Guiet, @nKiaru, Romain F. Laine for the feedback and sorry if I missed anyone!


Romain F. Laine introduces ZeroCostDL4Mic at the The Oslo NorMIC imaging node image processing course
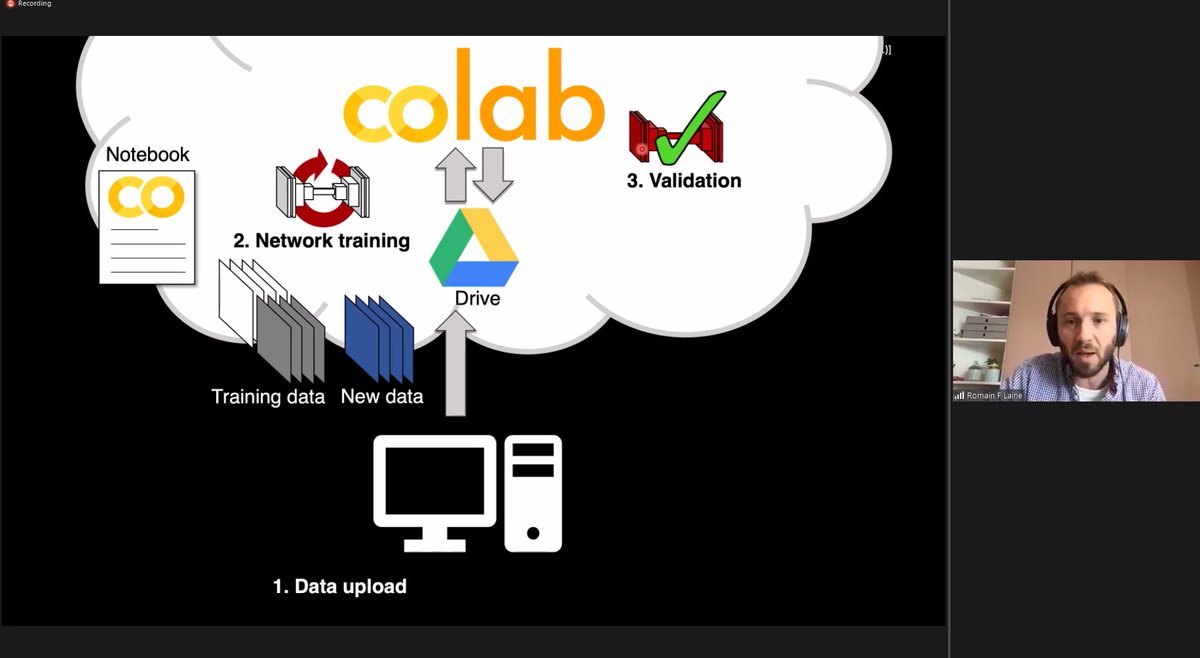

Playing with Ricardo Henriques Romain F. Laine Hannah Heil #eSRRF is fun 😁
biorxiv.org/content/10.110…
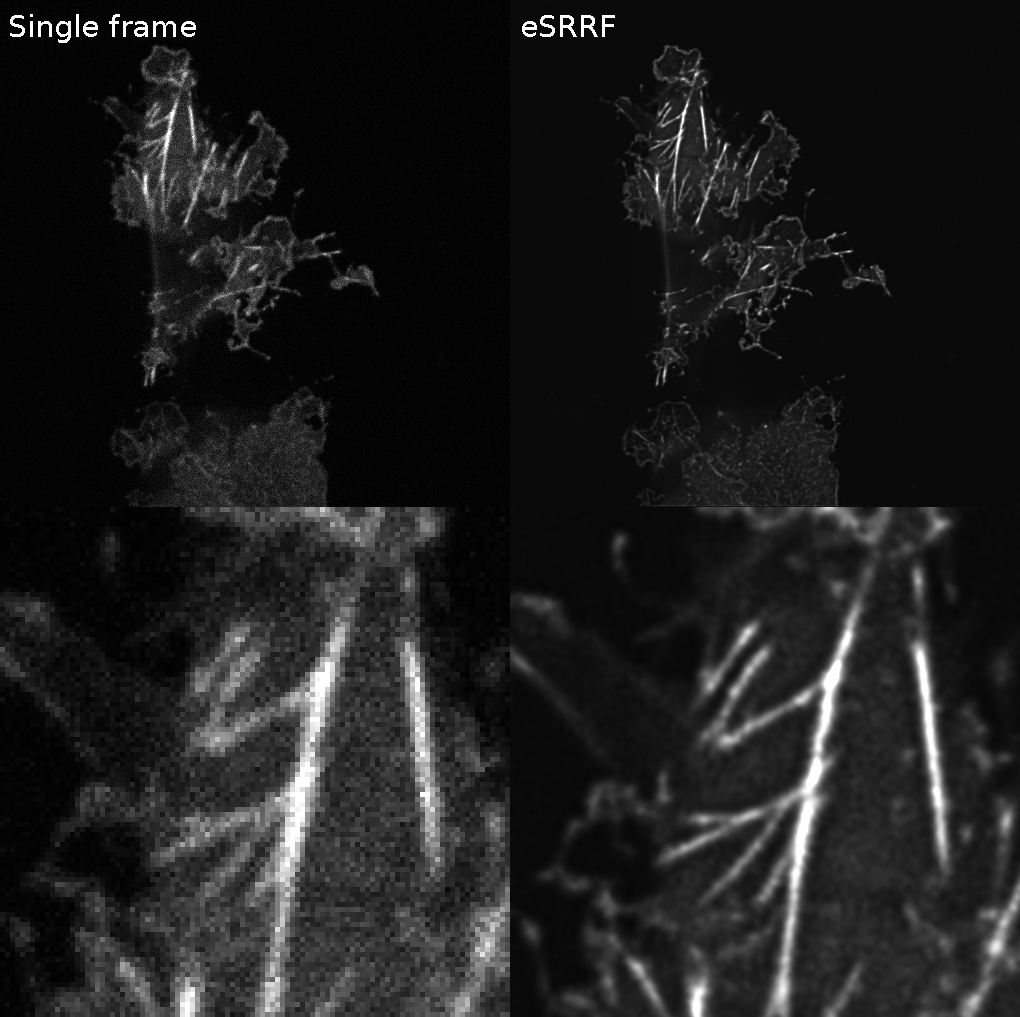
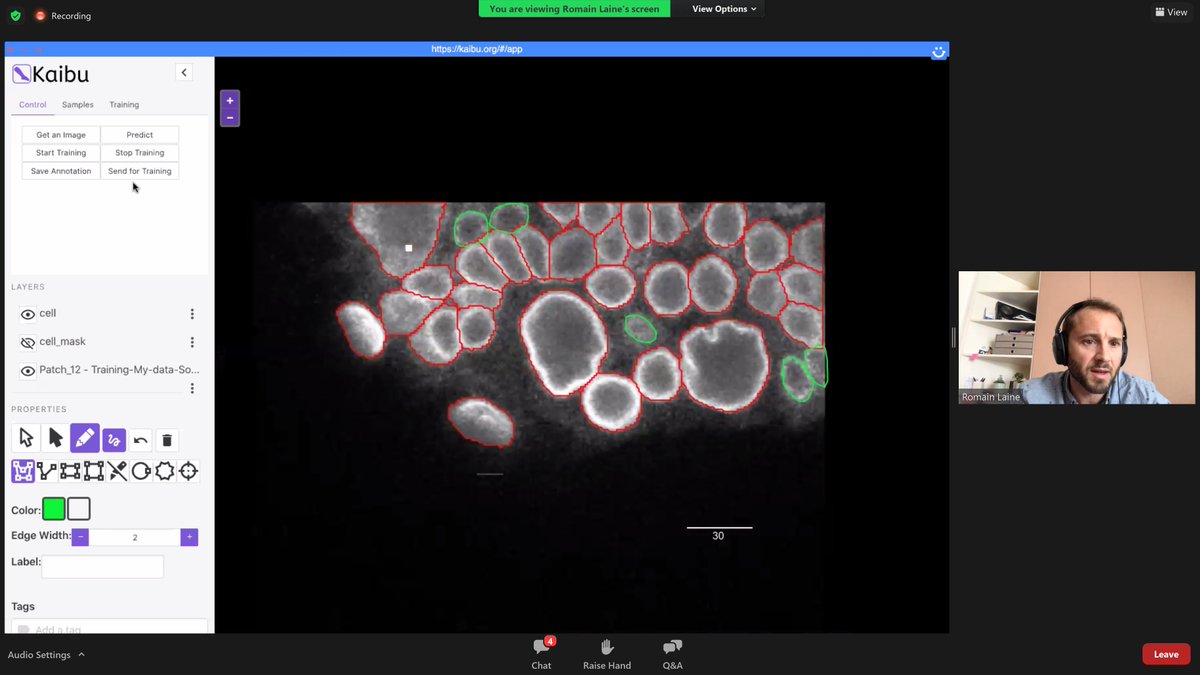
What a #NeubiasAcademy webinar! Guillaume Jacquemet and Romain F. Laine demo-ing #ZeroCostDL4Mic . Including applications integrating #TrackMate and #CellPose . Just now: interactively building a training dataset using #Kaibu .🤩🤯 @NEUBIAS_COST

But what is the *optimal* PSF-pair for the dense case? We used deep learning to find an optimal phase mask pair. The result is this “Nebulae” PSF pair (thanks Romain F. Laine for the name!). Information is encoded in the PSF shapes *and* in the relative lateral position (10/11)

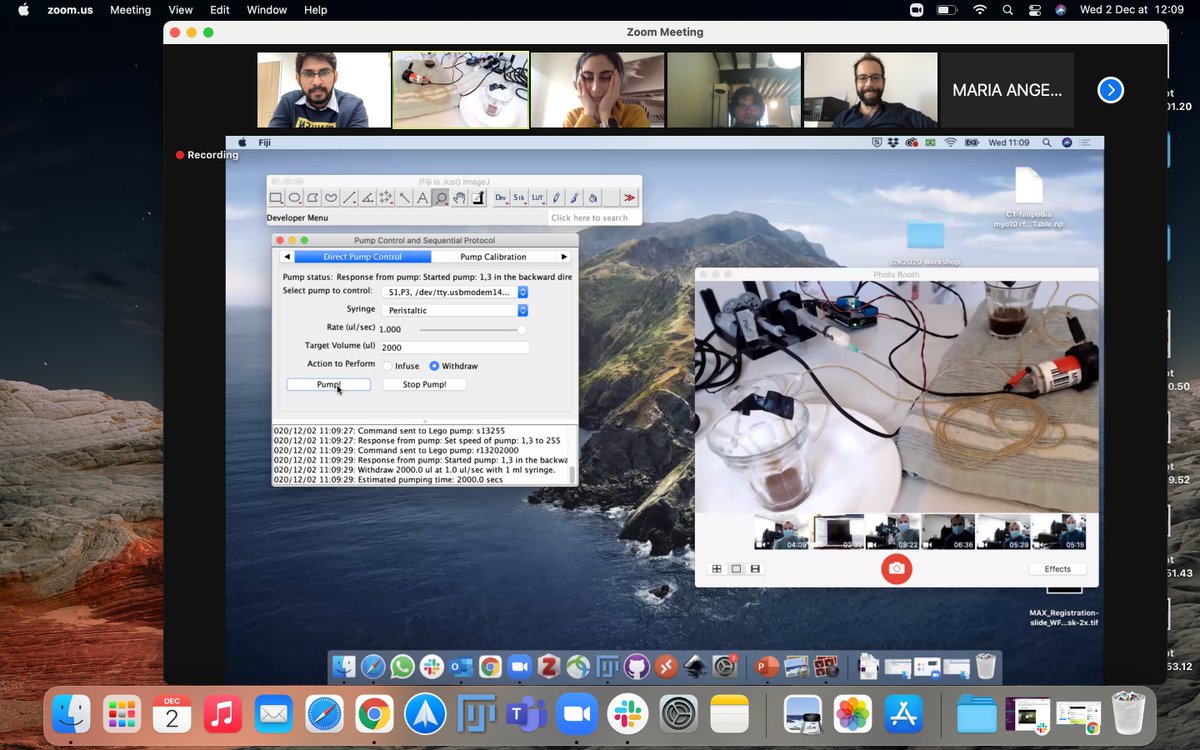
#i2k2020 has been by far the most interesting workshop. Very cool 'Pumpy' setup for sequential & automated labeling of samples - demonstrated by Romain F. Laine.
Cheap & resourceful!
Amazing tutorial Romain F. Laine Ricardo Henriques

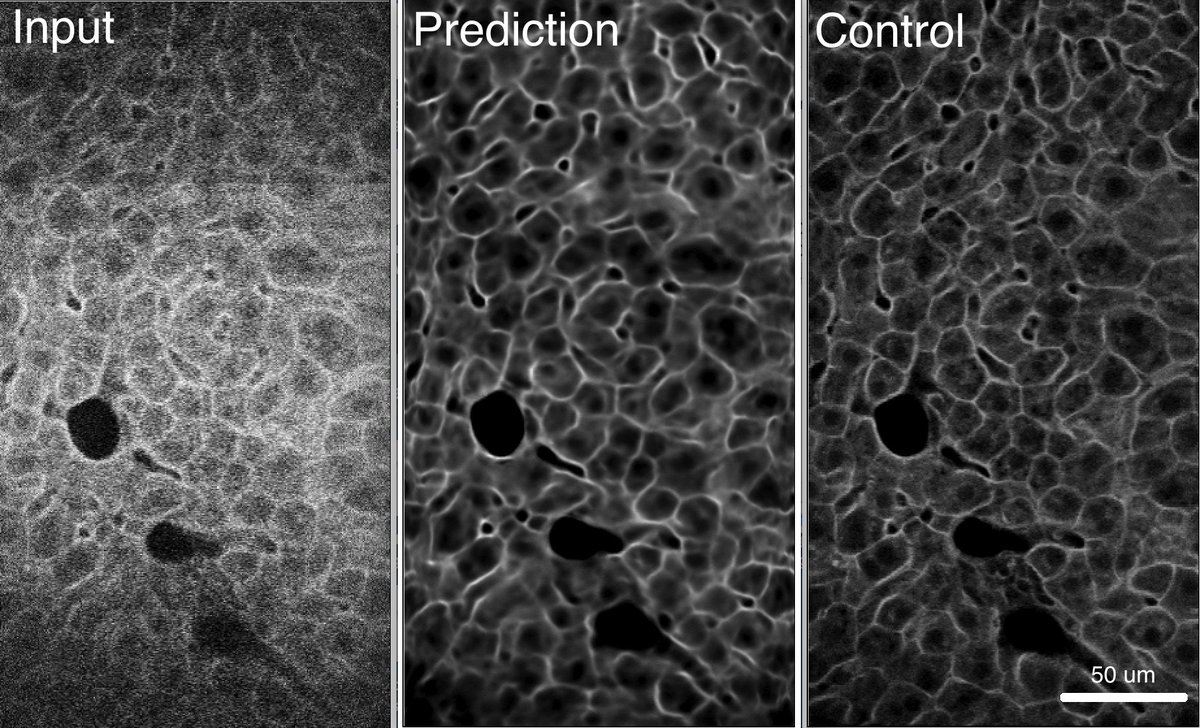
I had fun denoising high-speed 2p images (at 500 fps) via deep learning ( #CARE ) by Martin Weigert et. al. and made extremely easy to use through the cloud-based platform #ZeroCostDL4Mic by Lucas von Chamier Johanna Jukkala Romain F. Laine et. al. from Guillaume Jacquemet Ricardo Henriques

I wasn't really sure it was possible to automate alignment of vCLEM datasets, but Daniel Krentzel Martin Jones Matouš Elphick MC Ricardo Henriques Romain F. Laine & Chris Peddie made it happen! In one click!! In #napari 🌟🔬
biorxiv.org/content/10.110…
The Crick CZI Science
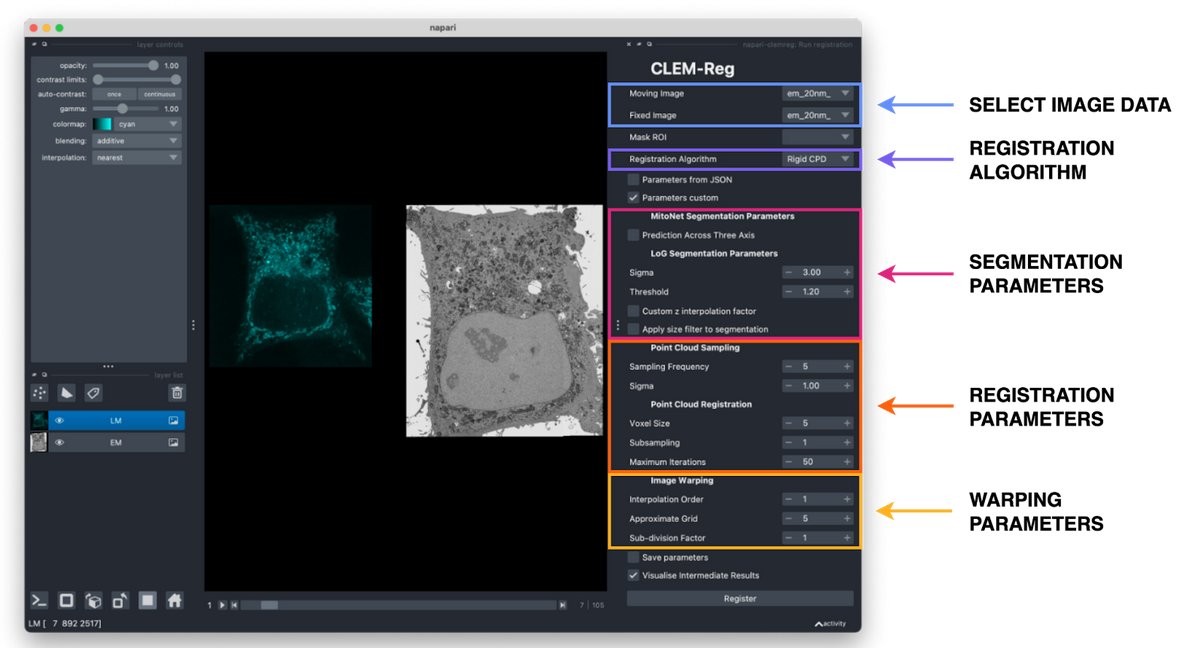

Here comes the next #I2K2020 tutorial highlight
In the #NanoJ hands-on tutorial with Romain F. Laine Lucas von Chamier Guillaume Jacquemet & Ricardo Henriques you'll learn tools to analyse #SuperResolution data while harnessing the power of your GPU
Janelia Conferences Pavel Tomancak @herrsaalfeld

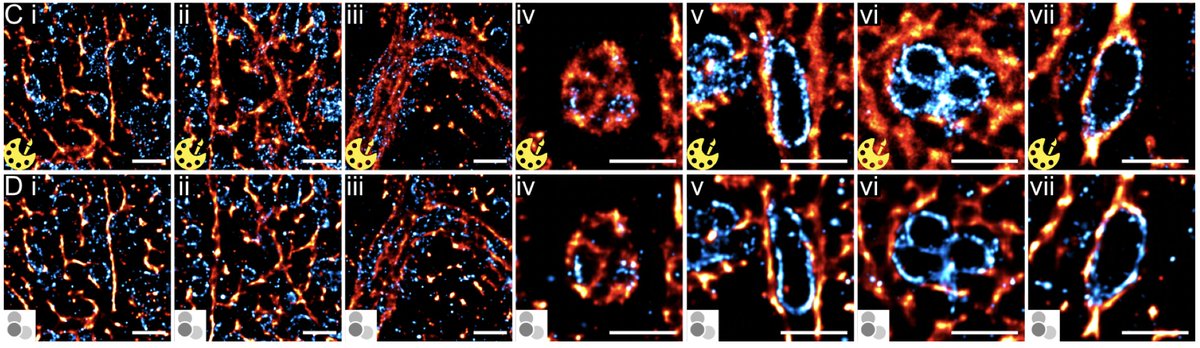
We employed the DeepSTORM NN developed by Nano Bio Optics Lab (Yoav Shechtman) and its implementation into the fantastic #ZeroCostDL4Mic platform Ricardo Henriques Guillaume Jacquemet Romain F. Laine to generate multi-color super-res images. Big thanks to Christoph Spahn and Elias Nehme (Shechtman Lab) for help!

For our #Microscopy data analysis course we will be joined by Romain F. Laine and Ricardo Henriques who recently published the #ZeroCostDL4Mic paper on #DeepLearning for #Microscopy paper in Nature. Read more below.
Course applications close 14 May - ebi.ac.uk/training/event…


Other new lessons include Maximum-Likelihood and EM algos as applied to superresolution microscopy—we're using a nice open-access dSTORM dataset of glial cells created by Christophe Leterrier & Romain F. Laine

Next @NeubiasAcademy webinar on June 15th at 15h30-17h00 CEST:
ZeroCostDL4Mic: exploiting Google Colab to develop a free and open-source toolbox for Deep-Learning in microscopy with Guillaume Jacquemet and Romain F. Laine.
Register here:⏬⏬⏬
tinyurl.com/n9562t9v