George Bouras
@gb13faithless
Bioinformatics @UniofAdelaide - phages, microbes and more
ID: 949343665
https://github.com/gbouras13 15-11-2012 08:44:29
2,2K Tweet
910 Followers
895 Following


🚨 Call for Abstracts! 🚨 One Health Aotearoa invites submissions for the #OneHealth Aotearoa Symposium 2024! 📆 3-4 December 📍University of University of Otago Submit your 250-word abstract by 31 August 2024 to [email protected] Don't miss out! #Research #Collaboration


I finally got around to testing HERRO (github.com/lbcb-sci/herro), the new Oxford Nanopore read correction algorithm which takes simplex ONT reads to PacBio-HiFi-level accuracy. I was impressed! Findings are on my blog in a two-part post: rrwick.github.io/2024/07/26/her… rrwick.github.io/2024/07/31/her…

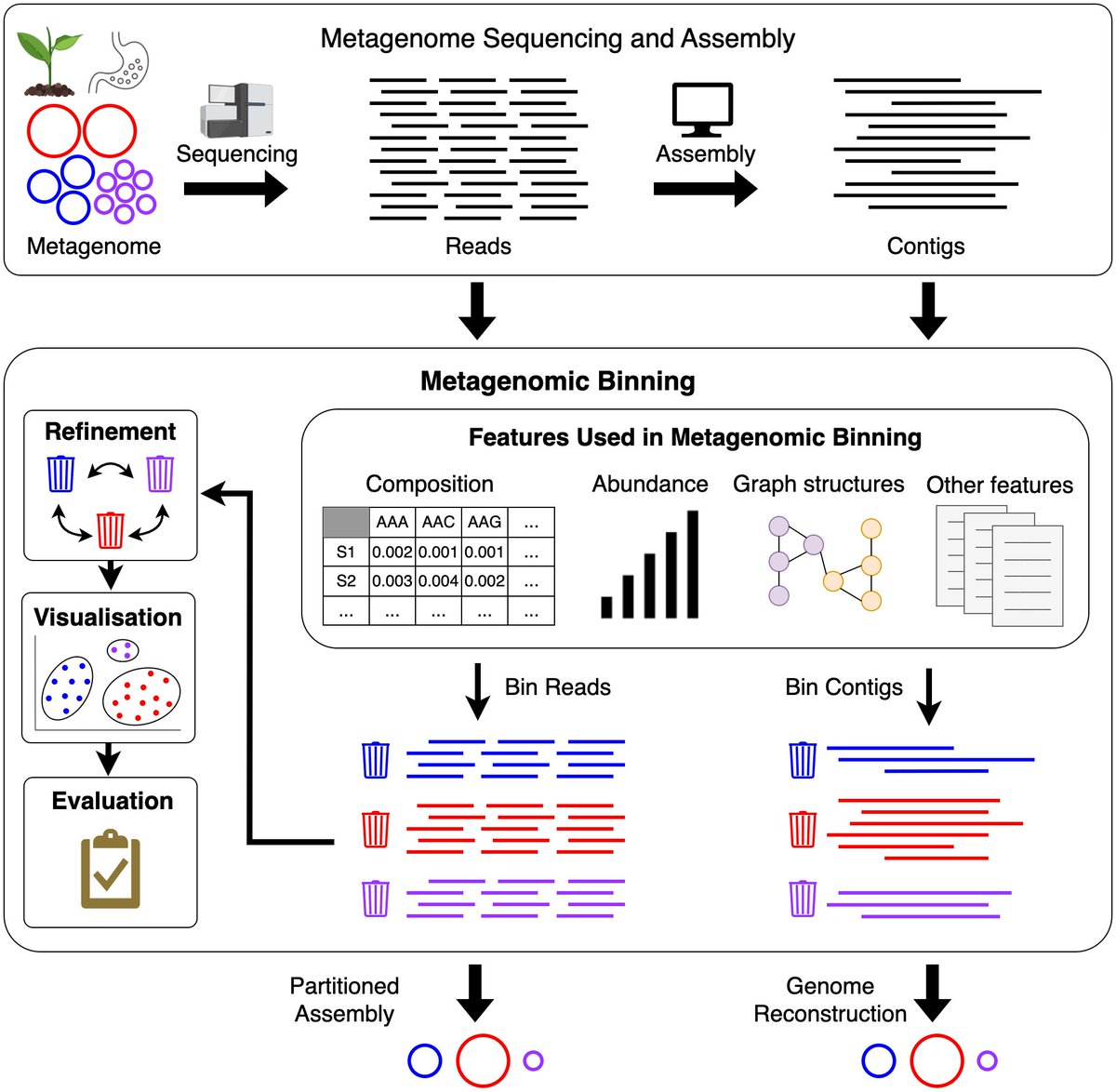
🧬 Excited to share our latest review article on metagenomic binning methods. (1/3) 👏 Kudos to co-authors Anuradha Hansheng Xue Bhavya Papudeshi Susie Grigson George Bouras DinsdaleLab Rob Edwards #Bioinformatics #Metagenomics #Binning #Software academic.oup.com/bib/article/25…

FoldMason progressively aligns thousands of protein structures in seconds, enabling remote MSA for distant phylogeny. Highlights: structural flexible MSA, LDDT conservation score, friendly webserver🧵 💾github.com/steineggerlab/… 🌐search.foldseek.com/foldmason 📄biorxiv.org/content/10.110…








Foldmason, our fast and accurate protein structure-base multiple sequence alignment method, has a logo thanks to HBKgenomics. Also, we updated the preprint, including a multi-domain protein benchmark. 📄biorxiv.org/content/10.110… 💾foldmason.steineggerlab.com 🌐search.foldseek.com/foldmason



We were lucky enough to hear Jon Mifsud present this work at #MicroSeq2024 yesterday. Published in Nature today! #Virology #Flaviviridae Check it out now👇 nature.com/articles/s4158…

Reconstructing virus evolutionary history is hard! Structural prediction and phylogenetics can help👍 Check out our new study out in nature nature.com/articles/s4158… University of Sydney Sydney Health

💻 Don’t miss George Bouras presenting Hybracter, a fast, automatic, scalable tool for recovering accurate bacterial genomes! Learn how it outperforms existing methods in recovering complete genomes and small plasmids. #MicroSeq2024 #Genomics #Bioinformatics


💻 Tune in to Bhavya Papudeshi’s presentation on Sphae, a toolkit for identifying #PhageTherapy candidates from #sequencing data! Learn how this workflow streamlines phage characterization for therapeutic use. #MicroSeq2024 #Bioinformatics