
François Simon
@francois__simon
Post-doc, Polytechnique Montreal.
Developped ExTrack, a tool to analyse single-particle tracking data. doi.org/10.1083/jcb.20…
ID: 1547578104859635714
14-07-2022 13:46:03
18 Tweet
32 Followers
105 Following

New #TrackMate extension for track analysis and kinetics studies: **TrackMate-ExTrack**. It is a port in TrackMate of the recent work by us (mainly @S_vanTeeffelen and François Simon) best escribed by Sven here: x.com/S_vanTeeffelen…

Video shows msfGFP-PBP1b single-particle tracking. François Simon, Jean-Yves Tinevez & @S_vanTeeffelen Université de Montréal present ExTrack, a tool to characterize single molecules from single-particle tracking bit.ly/3YaScfS #Biophysics #Membrane



Congratulations to the one and only Alexandra Colin ! The #CytoMorphoLab is happy (and proud) to count a new great member. Hold on the comet, she gonna fly high !



TARDIS (Temporal Analysis of Relative Distances) is now out in Nature Methods! nature.com/articles/s4159… It redefines single-particle tracking: spt is no longer limited by tracking algorithms - TARDIS offers at least 10x higher throughput, exceptional noise robustness: a 🧵


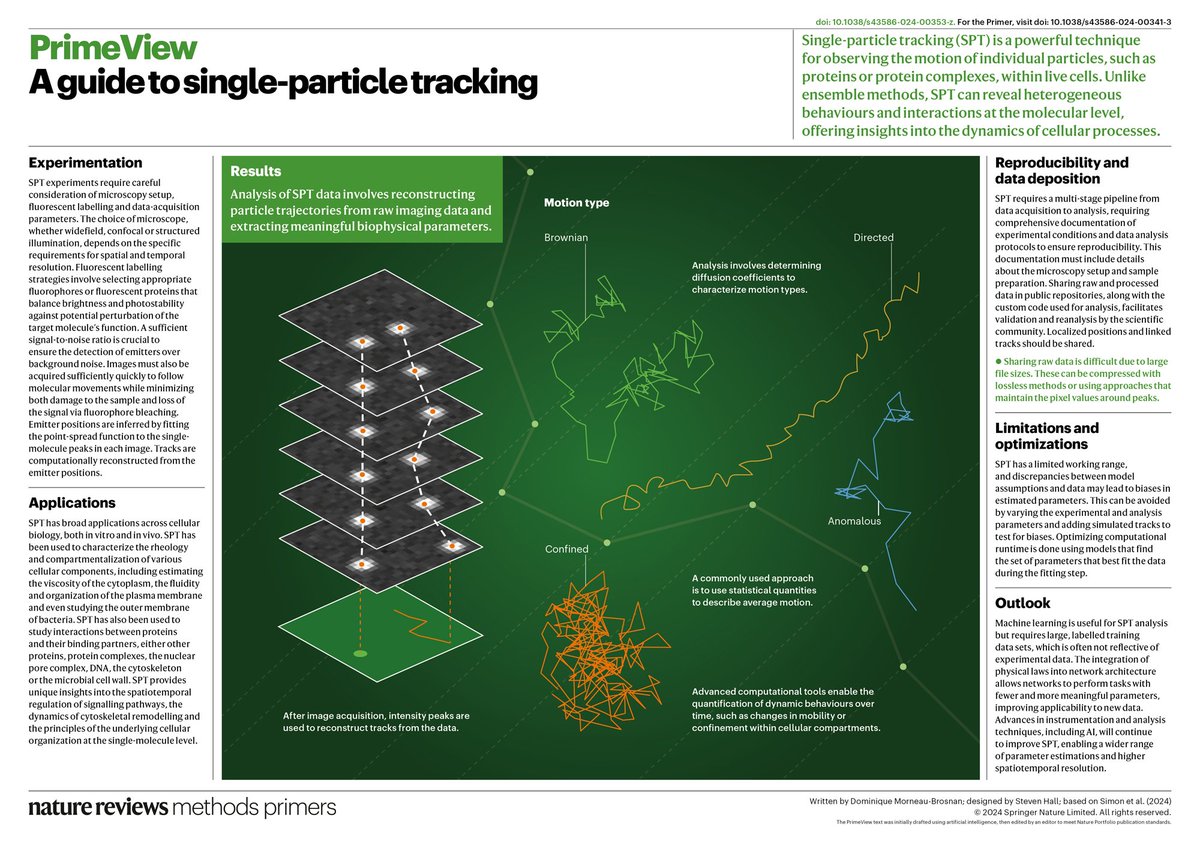
Single-particle tracking is a method for analyzing motion behaviors of individual proteins and complexes in live cells Check out our latest Primer to find out more: go.nature.com/3B033Dv Lucien Weiss François Simon @S_vanTeeffelen Faculté de médecine Université de Montréal Polytechnique Mtl


Woohoo! "A guide to single particle tracking" is now online! Together with François Simon & @S_vanTeeffelen, we share our thoughts on going beyond MSD analysis in Nature Reviews Methods Primers. doi.org/10.1038/s43586… Free read link: rdcu.be/dTKHd

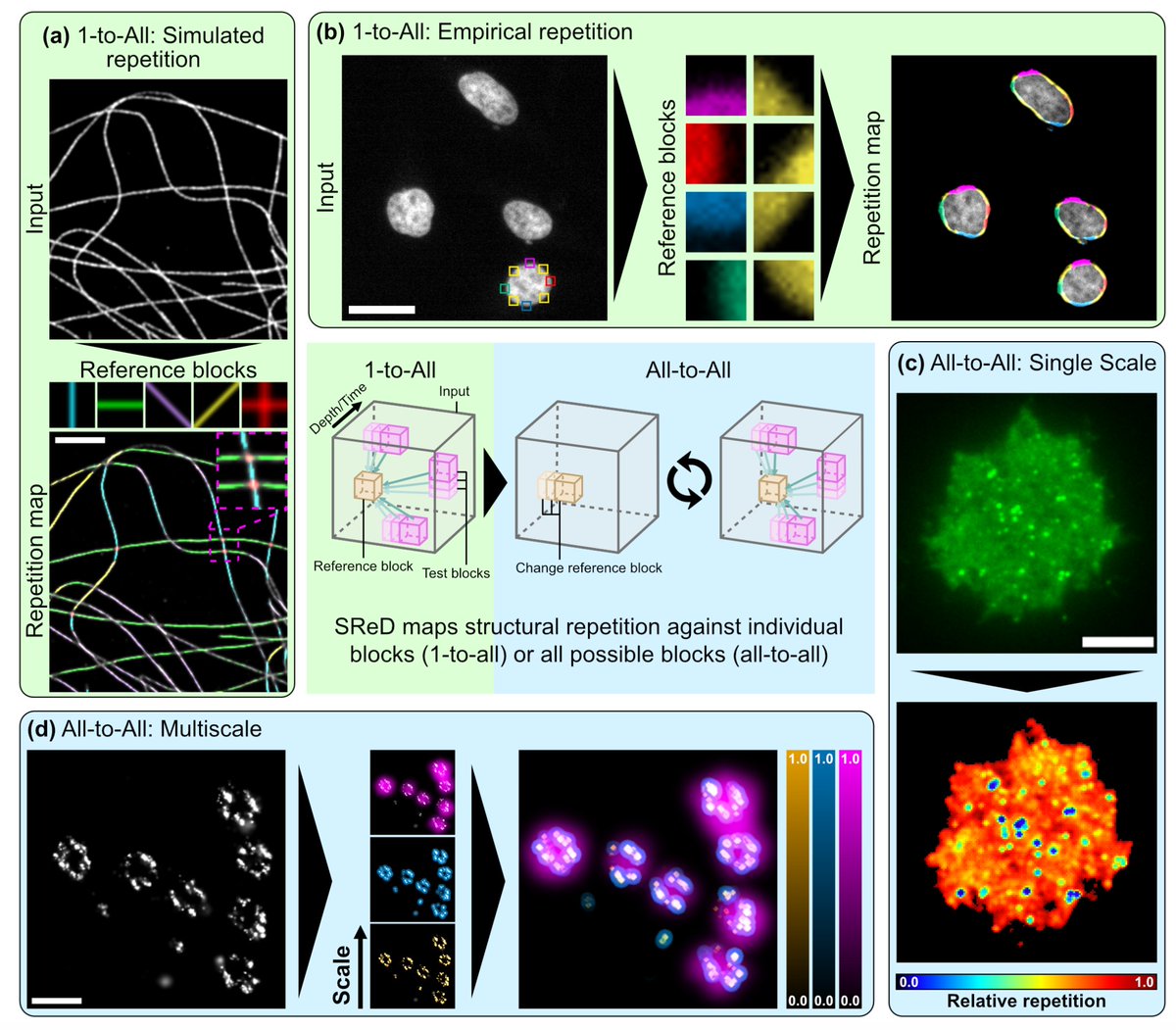
🚀🔬🦠 #SReD landed on bioRxiv! Detecting repetitive structures in #microscopy data - from neuron scaffolds to viral morphogenesis. GPU-accelerated multiscale pattern mapping in 💻Fiji! With Bruno Saraiva J Mamede @guijacquemet Christophe Leterrier biorxiv.org/content/10.110…

Hamidreza Heydarian Teun Huijben We also realised that quantitatively analysing structures without prior knowledge was not just OUR problem. So, we sought to demonstrate SReD's applicability in different experimental contexts, and release it as an #opensource package tailored for biologists!

I am more than happy to share our preprint about the evolution of non-visual and visual opsins in fishes, fruit of a collaboration between the Salzburger lab (Lily Fogg , Walter Salzburger Lab ) and Fabio Cortesi (Fabio Cortesi). biorxiv.org/content/10.110…