Lotfi Slim
@epislim
Solution Architect @Nvidia- AI/ML & Genomics
ID: 836293875052478467
27-02-2017 19:16:12
249 Tweet
178 Followers
1,1K Following


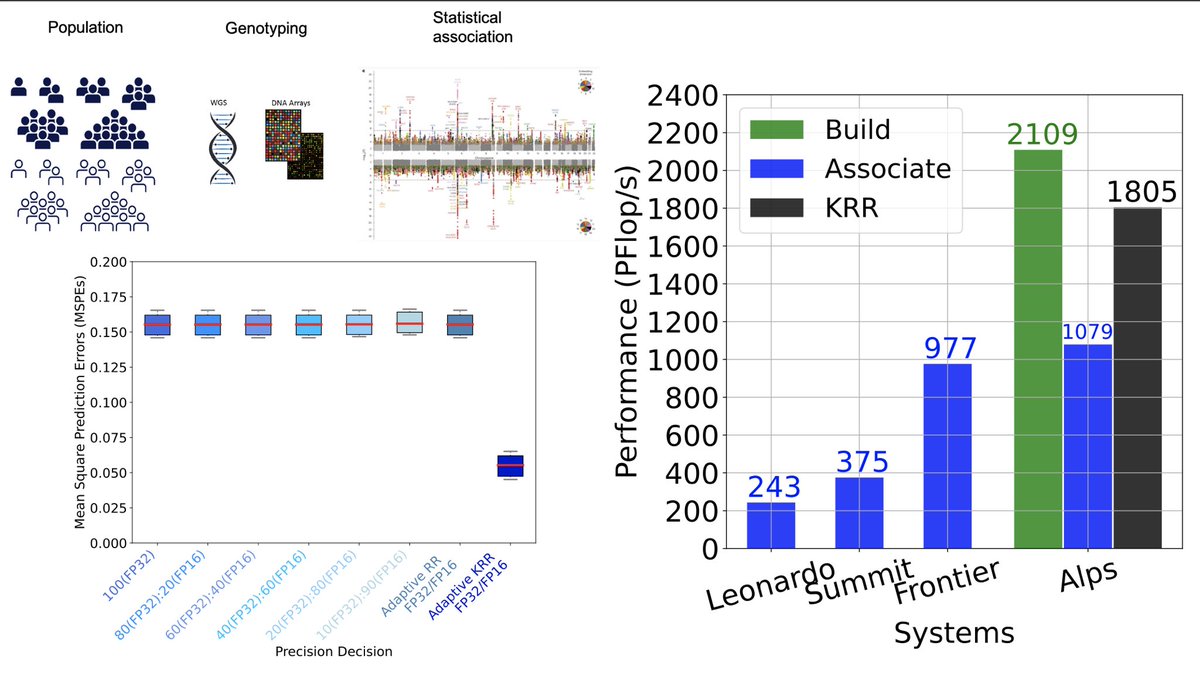
Epic with #genomics! Thanks Lotfi Slim and Rabab Alomairy, Ph.D. for this cool collaboration. Linear algebra comes again to rescue and addresses multiple precisions during genome-wide association studies. #GWAS Do not oversolve! Ask questions live on Mar 23 at 2pm CET. #HPC #GTC23



Great work from Hassan Sirelkhatim and Christian Dallago as always!









📣 When #exascale meets #bioinformatics #GWAS to capture nonlinear genetic #epistasis using #NVIDIA #MxP #GPU: - 1.8 EFlop/s on #Alps CSCS Lugano - 305K patients of UK #BioBank - 13M synth cohort, ~63% of the world's countries 🌍 - 10000X vs SOTA #REGENIE #GordonBell #SC24 #HPC