Zhana Duren都仁扎那
@durenzhana
Associate Professor at the Indiana University School of Medicine
ID: 1332087767812239360
https://durenlab.com/ 26-11-2020 22:24:21
42 Tweet
219 Takipçi
198 Takip Edilen










Call for Papers and Abstracts The 12th International Conference on Intelligent Biology and Medicine (ICIBM 2024) will be held in Houston, TX, USA. Please contact our main organizers (PC chairs) Qianqian Song @ecique_ or [email protected].



Zhana Duren都仁扎那 Clemson Center for Human Genetics Clemson University College of Science Clemson University Clemson researchers pave the way for precision medicine with AI #NBTintheNews via Clemson University news.clemson.edu/clemson-resear…



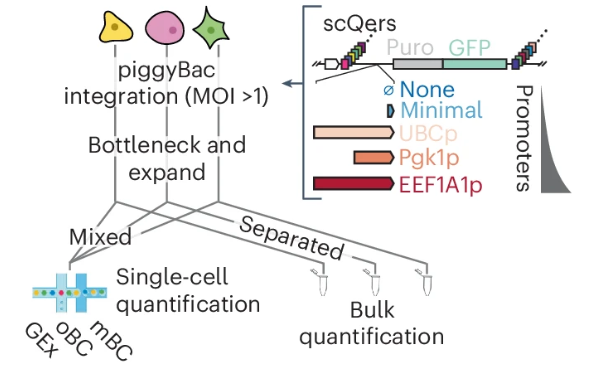
scQers are quantitative single cell expression reporters reported by Jay Shendure and colleagues. They enable high-sensitivity quantitative characterization of developmental cis-regulatory elements at the single-cell level. nature.com/articles/s4159…