
Dehui Lin
@dehuilin
Research officer at GIS A*STAR
ID: 1514617311218470926
14-04-2022 14:51:49
6 Tweet
23 Followers
87 Following

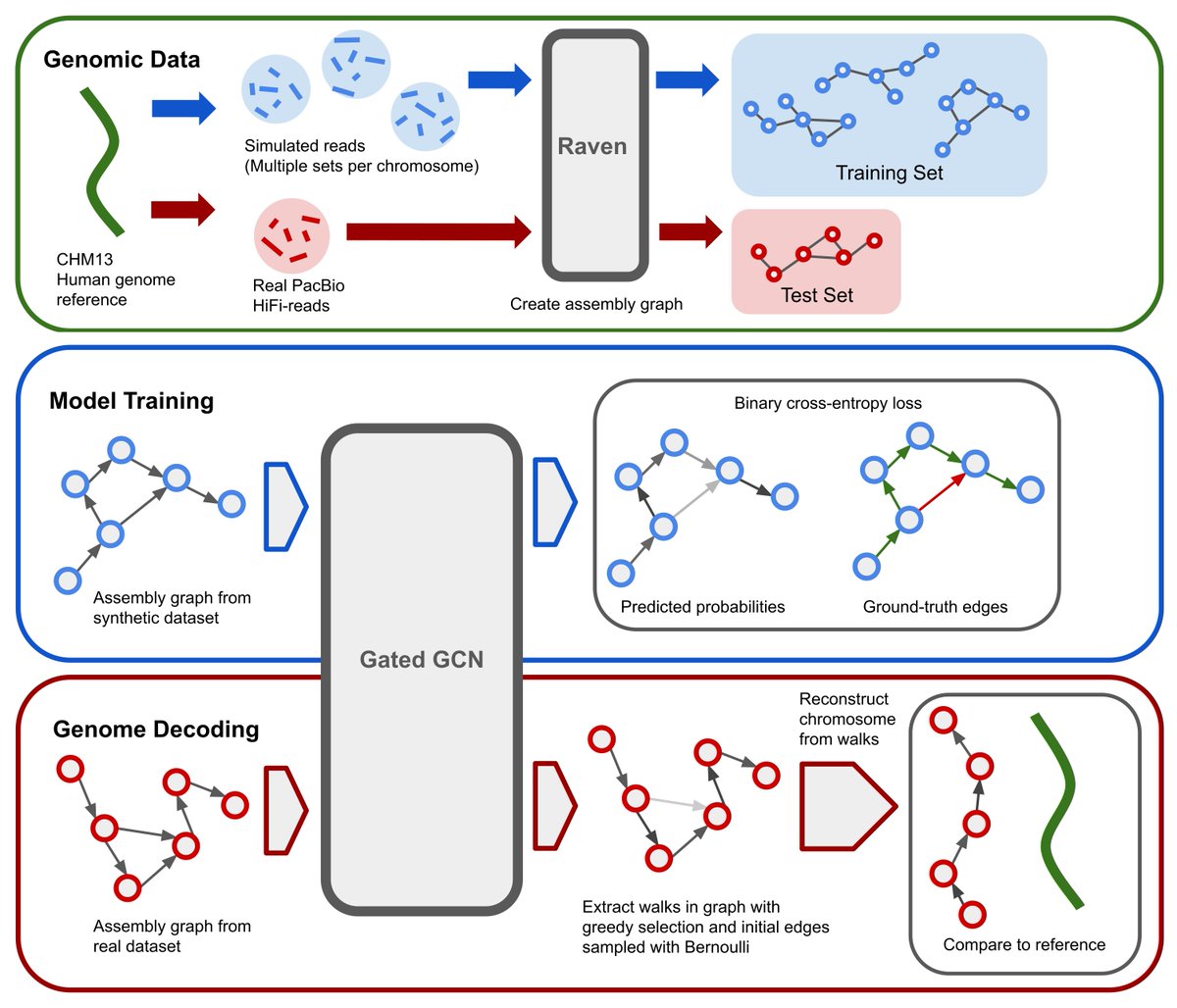
After many months, I'm proud to share our work on untangling genome assembly graphs with GNNs. Or, GNNome Assembly (sorry not sorry)🧬 Paper: arxiv.org/abs/2206.00668 Code/Data: github.com/lvrcek/GNNome-… with Xavier Bresson, T. Laurent, Martin Schmitz, and Mile Sikic. Read more in 🧵

Long reads metagenome benchmark is out. Highlights. 1. In most cases Kraken, 2. minimap2/ram for slightly higher accuracy. 3. The right database is of huge importance 4. Check taxonomy files carefully. bmcbioinformatics.biomedcentral.com/articles/10.11… w/Niranjan Nagarajan Krešimir Križanović @jmaricb Sylvain

Are nanopore UL reads only long reads we need? We developed Herro github.com/lbcb-sci/herro AI error correction model that can correct reads to accuracy above Q30 while trying to keep informative positions intact. w/ Dominik Stanojevic Dehui Lin Sergey Nurk 🇺🇦 Pore_XX_Singapore Oxford Nanopore



HERRO - imagine what you can do with accurate ultra-long nanopore reads biorxiv.org/content/10.110… w/ Dominik Stanojevic Dehui Lin Pore_XX_Singapore and great help from Sergey Nurk 🇺🇦 #nanopore. See you at London Calling 2024!!