
Deep Seq
@deepseqnotts
Core Next Generation Sequencing facility at the University of Nottingham. Experts in Nanopore, Illumina and Sanger sequencing
ID: 742969975
http://www.nottingham.ac.uk/deepseq/ 07-08-2012 14:02:46
161 Tweet
669 Takipçi
101 Takip Edilen

Just shy of 800 sequences contributed to COVID-19 Genomics UK (COG-UK) Consortium by our team since this all began. Thanks to all involved from UoN Life Sciences and Nottingham University Hospitals




About to start out 100th Oxford Nanopore run for covid samples with the team from Deep Seq . A sad milestone but amazing work by our local team including many from @TeamNUH and Patrick McClure all in support of the work of COVID-19 Genomics UK (COG-UK) Consortium .




So - our ReadFish paper came out on Monday nature.com/articles/s4158… using GPU - but what if you only have a laptop? Here's an update from alex to our tools that enable ReadFish on Mac, Linux or Windows with no additional GPU or compute. looselab.github.io/2020/12/02/rea… for details.



Well Oxford Nanopore - always seems to happen this time of year... bit of a bug in readfish but sure we will sort it out soon....

I still remember the first time I saw data from @scalene showing ultra long reads (N50>=100kb) were possible. Since then we and others (especially John Tyson) have worked to get longer reads. This weekend we tested the Circulomics Oxford Nanopore ultra long kits. Wow! 19.49 Gb N50 143kb
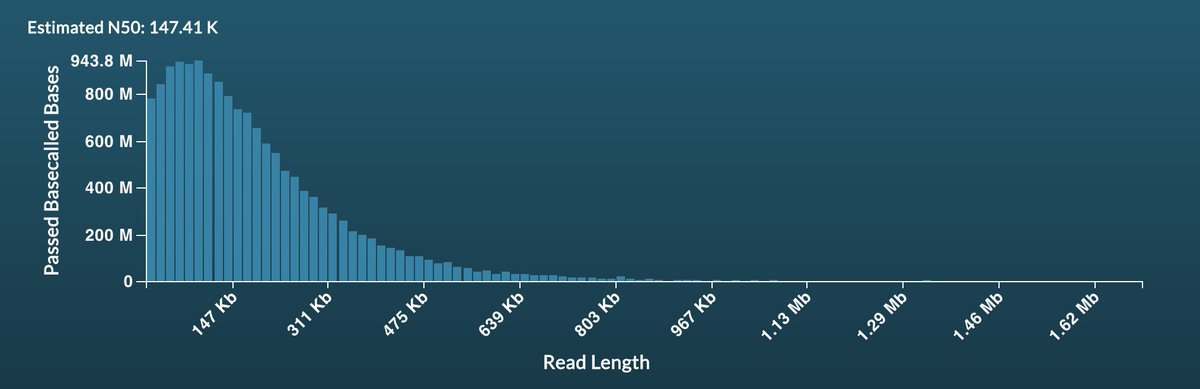

Earlier this week we put out a couple of preprints (doi.org/10.1101/2021.0… doi.org/10.1101/2021.0…) describing minoTour github.com/LooseLab/minot…, an Oxford Nanopore tool we have been developing for several years. A bit of a 🧵



This gif is from one sample of three run on a single Oxford Nanopore promethION flow cell. Frankly I'm super impressed at the performance though there is headroom for more improvements. We ran the same libraries as on gridION and have updated our preprint here. biorxiv.org/content/10.110…

Rapid #genome-wide copy number profiling using adaptive sampling on Oxford Nanopore by Matt Loose and the amazing team Deep Seq Real potential for clinical application.

We have an opportunity for someone to join our fabulous bioinformatics team - nottingham.ac.uk/jobs/currentva… - you will get to work with Oxford Nanopore Illumina @bionanogenomics 10x Genomics and more! Please retweet and dm for info!




