
Dheeraj Prakaash
@dj_biophys
Postdoc @SKhalidLab @UniofOxford | PhD @KalliGroup @UniversityLeeds @AstburyCentre | #CompBiochem #Tech #SciArt | Moved ➡️ dheerajprakaash.bsky.social
ID: 963315079767736320
https://www.linkedin.com/in/dheeraj-prakaash-b757679a/ 13-02-2018 07:33:07
678 Tweet
403 Takipçi
629 Takip Edilen






🚀 Check out our latest review, "Computational Methods for Modeling Lipid-Mediated Active Pharmaceutical Ingredient Delivery", now published in Molecular Pharmaceutics (ACS Publications )! 🧪📖 🔗 Read here: doi.org/10.1021/acs.mo…







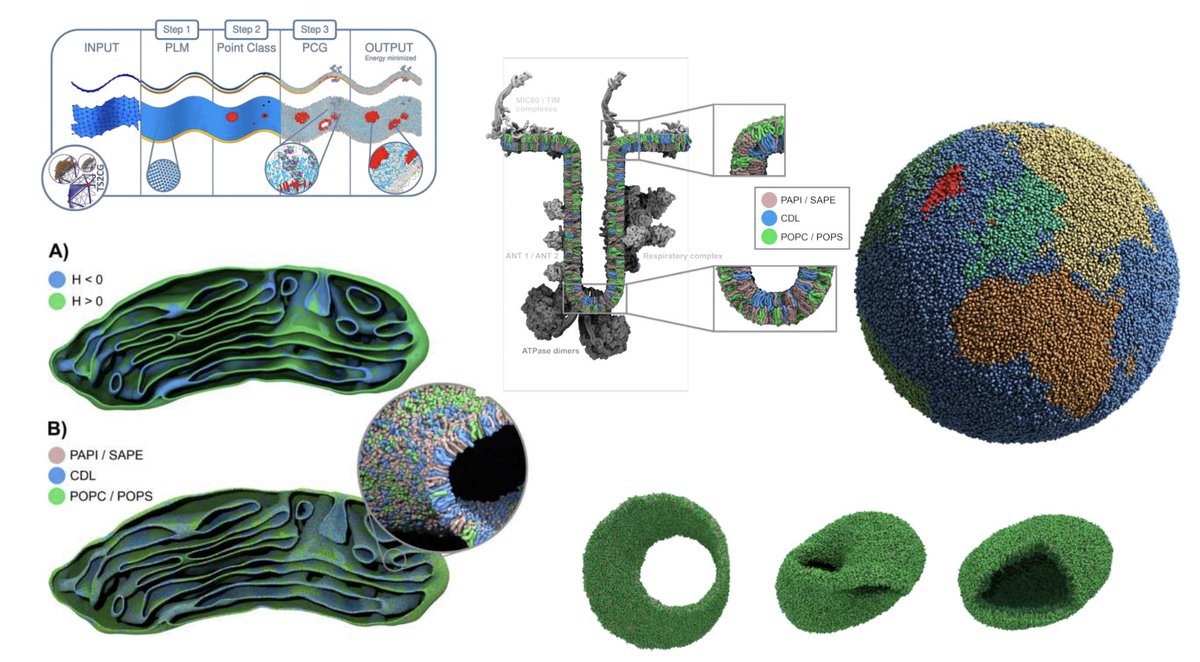
New preprint: TS2CG v2, building and simulating membranes with any shape and lateral organisation. We got Martini Globe, Martini Möbius strip and again mitochondrion. biorxiv.org/content/10.110… Siewert-Jan Marrink Source code: github.com/weria-pezeshki…







After a 4-year journey, we are super happy to see this paper out in Nature Communications - Katarina Elez et al: High-throughput molecular dynamics + active machinelearning enable efficient identification of an experimentally-validated broad coronavirus inhibitor. nature.com/articles/s4146…


