Conor Walker
@bioconorwalker
Bioinformatics Scientist.
Formerly postdoc at @nygenome/@Columbia and PhD @emblebi/@Cambridge_Uni.
ID: 844440568256221185
http://conorwalker.net 22-03-2017 06:48:15
58 Tweet
156 Takipçi
696 Takip Edilen

We developed a new method (phastSim) to simulate sequence evolution along huge phylogenies, in particular pandemic-scale ones like in SARS-CoV-2 doi.org/10.1101/2021.0… with Goldman Group Russ Corbett Conor Walker Lukas Weilguny Yatish Turakhia
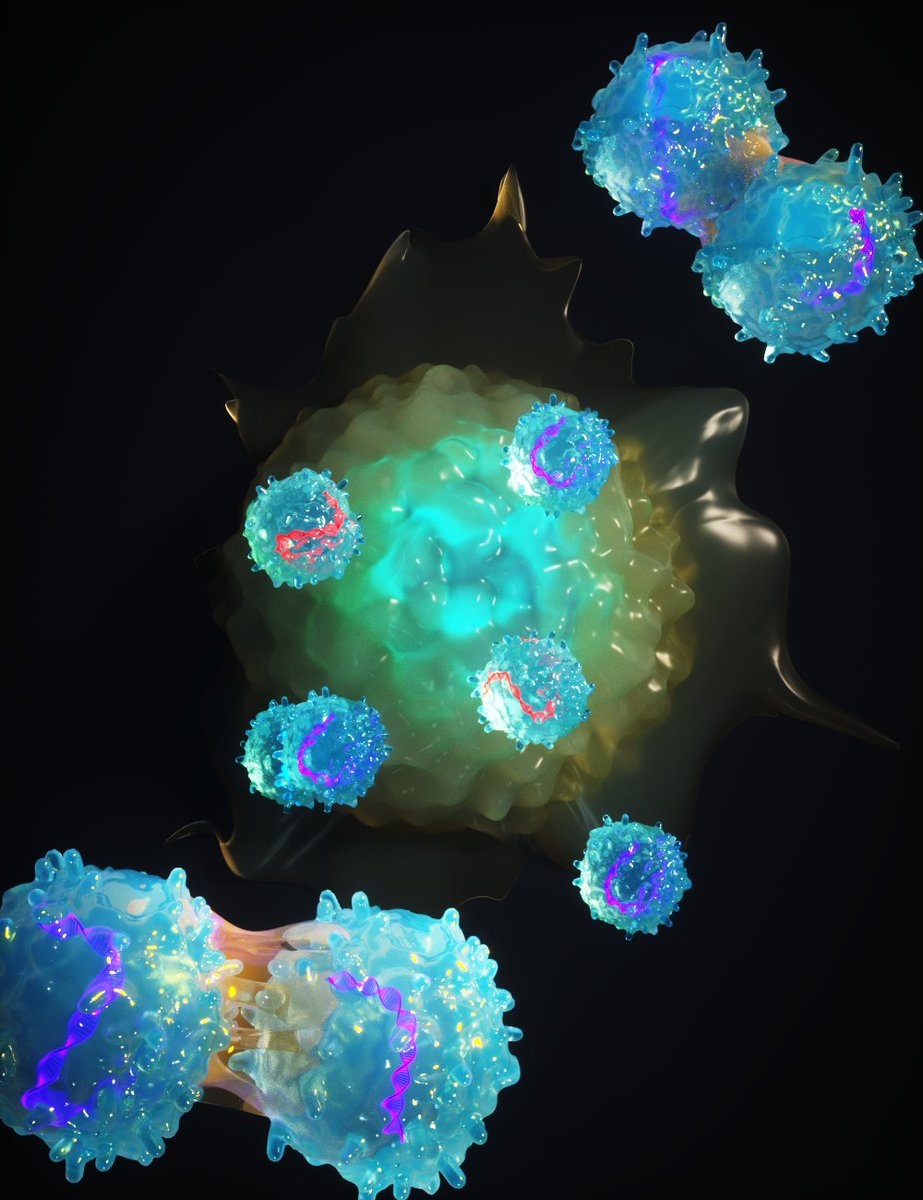
For those of you who love to explain complex mutation clusters parsimoniously (who doesn't?), the final version of Conor Walker's paper is now out at journals.plos.org/plosgenetics/a… - also feat. Aylwyn Scally and Nicola De Maio. And me. Cool figures too: this 👇 is part of S.Fig 10




The impressively huge scale of SARS-CoV-2 sequencing has really stretched our ability to do phylogenetics, and is leading to some fantastic advances in methods. Here's one that Nicola De Maio from the Goldman Group group at EMBL-EBI contributed to. x.com/emblebi/status…


Congrats to the 2021 EvolCompGen Talk & Poster awardees for #ismbeccb2021 Best talk - Elise Parey (Camille Berthelot @camilleberthelot.bsky.social lab) Best poster - Vignesh Sridhar (Vignesh Sridhar, JRaviLab) Honorable mentions Talks - Uno Punto c Conor Walker @matteodelabre Posters - Elliot Majlessi




Groundbreaking work from Nicola De Maio Goldman Group and colleagues "we rewrite the classical Felsenstein pruning algorithm so that we can infer phylogenetic trees on at least 10 times larger datasets with higher accuracy than existing maximum likelihood methods. "





If you were looking for evidence that isoforms are cool, all-pervasive in the 🧠, and everybody should study them at the single cell level, our work on mouse development and brain-region variability from the Hagen Tilgner / @[email protected] lab is now live Nature Neuroscience! 1/ nature.com/articles/s4159…

Super excited to share my lab's latest work on quantifying the private information leakage from single cell count matrices. Led by very first postdoc in the g2lab, brilliant Conor Walker, we showed that individuals can be reidentified 1/n cell.com/cell/fulltext/…

![Goldman Group (@ebigoldman) on Twitter photo Simulating pandemic-scale genome sequencing datasets, e.g to benchmark your new analysis methods? Try phastSim, now updated to include indels (biorxiv.org/content/10.110…)
[1/2] Simulating pandemic-scale genome sequencing datasets, e.g to benchmark your new analysis methods? Try phastSim, now updated to include indels (biorxiv.org/content/10.110…)
[1/2]](https://pbs.twimg.com/media/FADWocgXoAIStrE.jpg)