
backofenlab
@backofenlab
Bioinformatics Lab - Department of Computer Science - University of Freiburg - bioinf.uni-freiburg.de/impressum.html
ID: 822466775627530246
http://www.bioinf.uni-freiburg.de 20-01-2017 15:32:14
246 Tweet
419 Followers
103 Following

Project Jupyter notebooks running in #usegalaxy and analysing the result of the 3k+ #SARSCoV2 workflow. RStudio is also available of course :) Thanks Anton Nekrutenko 🇺🇦 for presenting and Daniel Blankenberg for his awesome work in Interactive Tools.


NanoGalaxy: Nanopore long-read sequencing data analysis in Galaxy. Our work on bridging Oxford Nanopore and Galaxy Project, the two accessibility heavens of sequencing & data analysis, is now published: doi.org/10.1093/gigasc… Wandering Wim @shiltemann Björn Grüning Marius van den Beek





Heartfelt congratulations to Milad Miladi for successfully defending his PhD thesis last friday. He published an amazing number of papers, 12 of them with a major contribution! Check out his work here: orcid.org/0000-0002-0173… #bioinformatics #COVID19 #bioconda #RNA #usegalaxy


Today Sergei Pond will talk about "Insights from selection analysis of complete genomes and read-level data". Join us and learn more about the genetics of #SARSCoV2. ELIXIR Europe #denbi #usegalaxy

We welcome our 1080 registered trainees! Don't hesitate to ask any of the 50+ trainers from all over the world. galaxyproject.eu/posts/2021/02/… ELIXIR Europe EOSC-Life #denbi Universität Freiburg Gallantries

🦠Don't miss out on the next #COVID19 Galaxy webinar with Björn Grüning @babi2305 and Wolfgang Maier With them, you will explore the Viral Beacon and Galaxy variant workflows ➡️bit.ly/GalaxyCOVID19 Centre for Genomic Regulation (CRG) Galaxy Project de.NBI / ELIXIR-DE The GalaxyP Project #usegalaxy Universität Freiburg


de.NBI / ELIXIR-DE and RBC Freiburg backofenlab will also support the next Galaxy Project Papercuts CoFest #usegalaxy #Bioinformatics. For more information see: denbi.de/training/1194-…



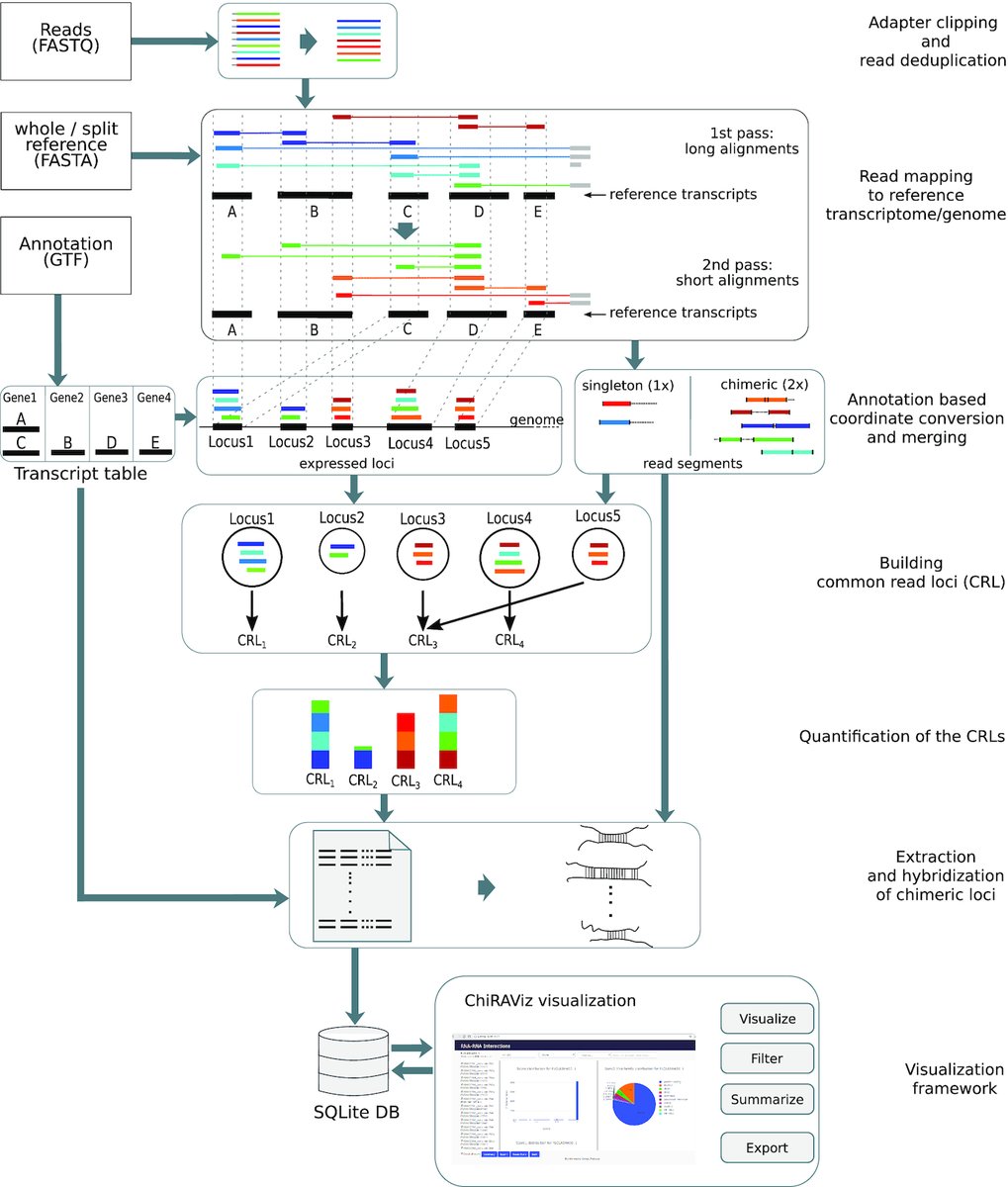
A new #usegalaxy tool suite from backofenlab for chimeric RNAs associated with specific cancer types. ChiRA: an integrated framework for chimeric read analysis from RNA-RNA interactome and RNA structurome data doi.org/10.1093/gigasc…

Are you interested in developing an online data analysis game in the context of Street Science Community? This student position is for you, apply now! #streetscience #CitizenScience #OpenScience


Exciting news, our preprint about predicting #ORF in bacteria from #Ribo-seq data is available. Great collab: Rick Gelhausen, @SarahLSvensson, Omer Alkhnbashi, Sharma Lab, @eggtron, backofenlab biorxiv.org/content/10.110… github.com/RickGelhausen/… #proteins #benchmark #Bioinformatics #SPP2002

Day #2 of the #DFG SPP2002 conference in Kiel. 🙂This morning Lydia Hadjeras, PhD from our group will present our joint Z-project with backofenlab on ribosome profiling and bioinformatics approaches for global sORF identification.