
Amjad Askary
@AskaryLab
Assistant Professor @UCLA |
Interested in using synthetic biology to better understand development and control cell fate | Views and opinions change over time
ID:3756492614
https://amjad.mcdb.ucla.edu/ 02-10-2015 07:15:49
522 Tweets
847 Followers
821 Following

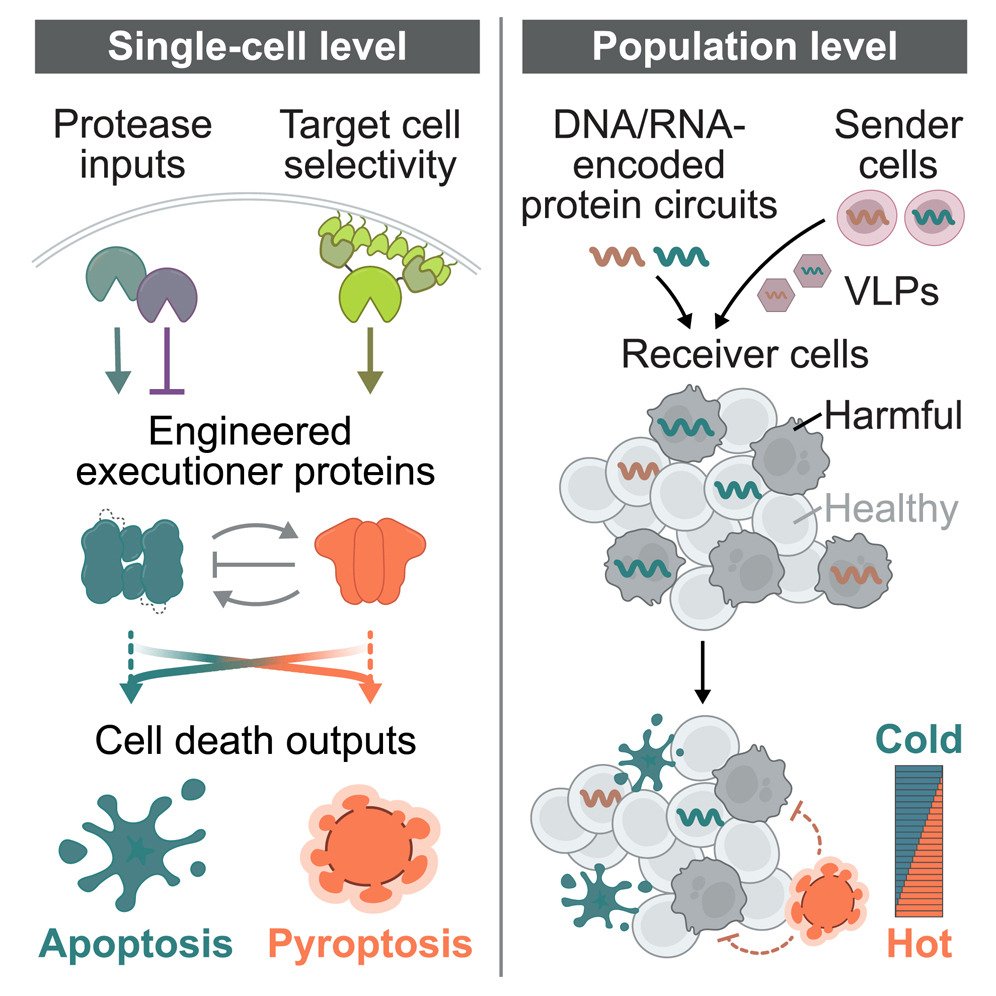
Our new paper on “synpoptosis” circuits, led by the amazing Shiyu Xia (夏时雨), is now out: sciencedirect.com/science/articl… (1/3)

As a side product, while developing SMORE, Zain Samadi also reimplemented the original STREME algorithm in MATLAB. This is especially convenient for Windows users: github.com/zsamadi/MATLAB…


Excited to have lineage motifs out in the world. Hoping the concept and methods can be useful in various developmental systems as more lineage data become available. Congratulations to Martin Tran and Amjad Askary on the work, and thanks to Adara Koivula and Inna-Marie for…






Our paper with Eric Deeds is now up on PLoS Comp Bio. Some new goodies since the preprint including an implementation of a generalized form of the branch distance!
journals.plos.org/ploscompbiol/a…


