
Melissa Haendel
@ontowonka
At the intersection of biology and everything else. #AttributionForAll. #UnitedAgainstHate #raredisease #FactsMatter, also at @[email protected]
ID: 777135943
24-08-2012 01:23:24
2,2K Tweet
2,2K Followers
2,2K Following

Happening now! Melissa Haendel presenting on behalf of N3C, one of the 2 Grand Prize Winners for FASEB DataWorks! We are honored and proud of our team at CTSA Data2Health #teamScience #openScience #realWorldData datascience.nih.gov/news/march-dat…


Next week, as part of National Human Genome Research Institute’s Genomic Innovator Seminar Series, Melissa Haendel and Jessica Chong will be speaking on data-driven approaches to defining rare genetic conditions. Register for the seminar, which takes place March 23 at 3 p.m. EST, here: allof-us.org/41C0K2b




With many kudos to justaddcoffee for leading this truly innovative work! Slides are here: bit.ly/amia23clusteri… Monarch Initiative HPO AMIA #IS23 #ontologies #realWorldData

How are researchers using genomic data to discover rare genetic conditions? At our Genomic Innovators Seminar, Jessica Chong and Melissa Haendel will discuss how researchers can use data to discover hundreds of genomic variants associated with rare diseases and improve disease diagnoses.


#Tislab Monarch Initiative is hiring! Assistant Research Professor in #Ontologies & Biocuration and a #Semantic Engineer: cu.taleo.net/careersection/… cu.taleo.net/careersection/… OBO Foundry Int Soc Biocuration OHDSI CTSA Data2Health AMIA

In our JC today, Vincent Strehlow 🗣️ about the 2022 ClinGen 📜 to guide ‘‘lumping & splitting’’ decisions. Thaxton et al. Melissa Haendel Heidi Rehm 4⃣ key criterias: assertion, molecular mechanism, phenotypic expressivity & inheritance pattern. 👇 sciencedirect.com/science/articl…



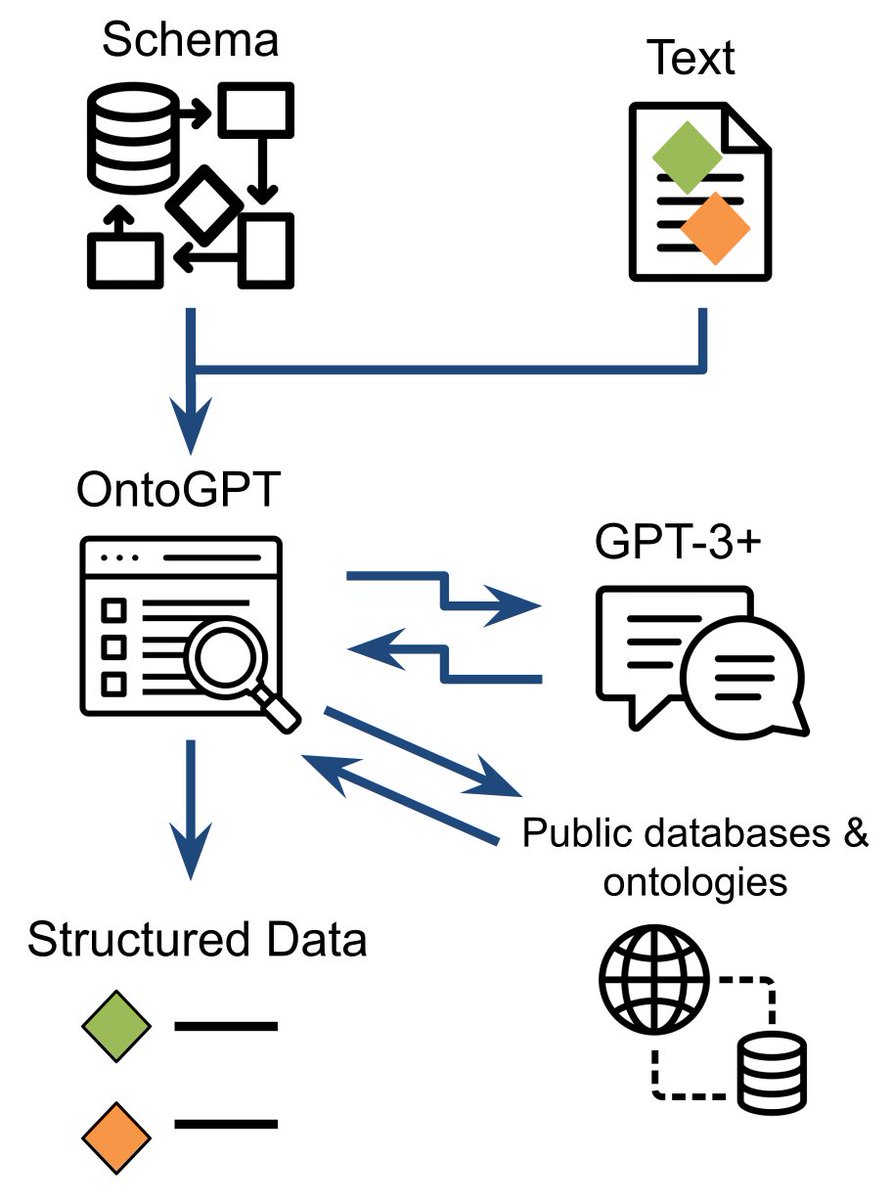
Here's our pre-print describing our GPT-3 based knowledge extraction tool SPIRES: arxiv.org/abs/2304.02711. Great work from J. Harry Caufield et al! SPIRES allows you to specify a knowledge schema (in LinkML - Linked (Open) Data Modeling Language) and then populate instances of that schema from unstructured text

We did it!! 💪🥳 This work was started during my PhD CU Anschutz Medical Campus and completed during my Postdoctoral Fellowship Columbia DBMI. There are so many important people to thank for making this work possible (please read all replies) 👇👇👇

Congrats! Our paper on De-black-boxing health #AI: demonstrating reproducible #machineLearning computable phenotypes using the N3C-RECOVER #LongCovid model in the AllofUsResearch data repository is out! NCATS CTSA Data2Health #N3C doi.org/10.1093/jamia/…


Honored to partner with our amazing team to enhance the AllofUsResearch program's data to advance #precisionmedicine & #realWorldData analytics! news.cuanschutz.edu/news-stories/n…

Harnessing consumer wearable #digitalBiomarkers for individualized recognition of #postpartumDepression using the AllofUsResearch dataset medrxiv.org/content/10.110… Congrats Eric Hurwitz!

"Elements of the [National COVID Cohort] collaborative’s technical approach will now be applied to AllofUsResearch, in the new center led by #N3C lead investigator Melissa Haendel, chief research informatics officer at the CU Anschutz Medical Campus."

Starting in a few mins: HL7 International GA4GH Monarch Initiative roundable on #phenotypic data exchange for #genetics & #raredisease. All are welcome! linkedin.com/posts/hl7-vulc…

Benchmarking #variantprioritzation tools for #raredisease diagnostics is critical! See Monarch Initiative new eval framework at: pubmed.ncbi.nlm.nih.gov/38915571/ and rebuttal of recent tool at: zenodo.org/records/127548…