
Jared Simpson
@jaredtsimpson
Computational Biologist. Principal Investigator at OICR & Assistant Professor @UofTCompSci.
ID: 74599038
http://simpsonlab.github.io/ 16-09-2009 00:05:45
2,2K Tweet
6,6K Followers
1,1K Following

An ultra-long read update. Since Circulomics is "no longer available" for Oxford Nanopore people might be interested in these protocols from longreadclub Ninin UoN LongRead and John Tyson - a toolkit of options here dx.doi.org/10.17504/proto… including monarch from New England Biolabs - 🧵below with more.



Join us at U of T Department of Computer Science! We are now hiring for tenure-stream faculty positions in multiple areas. We will start reviewing applications on December 6, and we will give full consideration to all applications submitted by January 10. Get the details: uoft.me/7jH



Me and Yun William Yu's work on local k-mer selection techniques is now accepted to Bioinformatics! We investigate both practically and theoretically the properties of different local k-mer selection techniques, the most well-known being the minimizer. academic.oup.com/bioinformatics…


Kar-Tong Tan found Oxford Nanopore basecallers often systematically call telomere repeats (TTAGGG)n into other repeats with similar signal profiles. He tuned the bonito basecaller and developed a pipeline to greatly reduce such errors. Read the thread and our preprint for details.



Join my team! Challenging but impactful projects, great colleagues, exciting science Ontario Institute for Cancer Research (OICR)! recruitingsite.com/csbsites/oicr/…


nanopolish v0.14.0 released, with support for modification bam files (thanks @BonfieldJames and Chris Wright) and compiling on M1 macs

Interested in software development for biomedical and imaging data? With Hartland Jackson we're looking for a developer for an exciting academic/industry collaboration including some world-unique highly multiplexed imaging data Details ➡️ apply.interfolio.com/106773 DMs open!

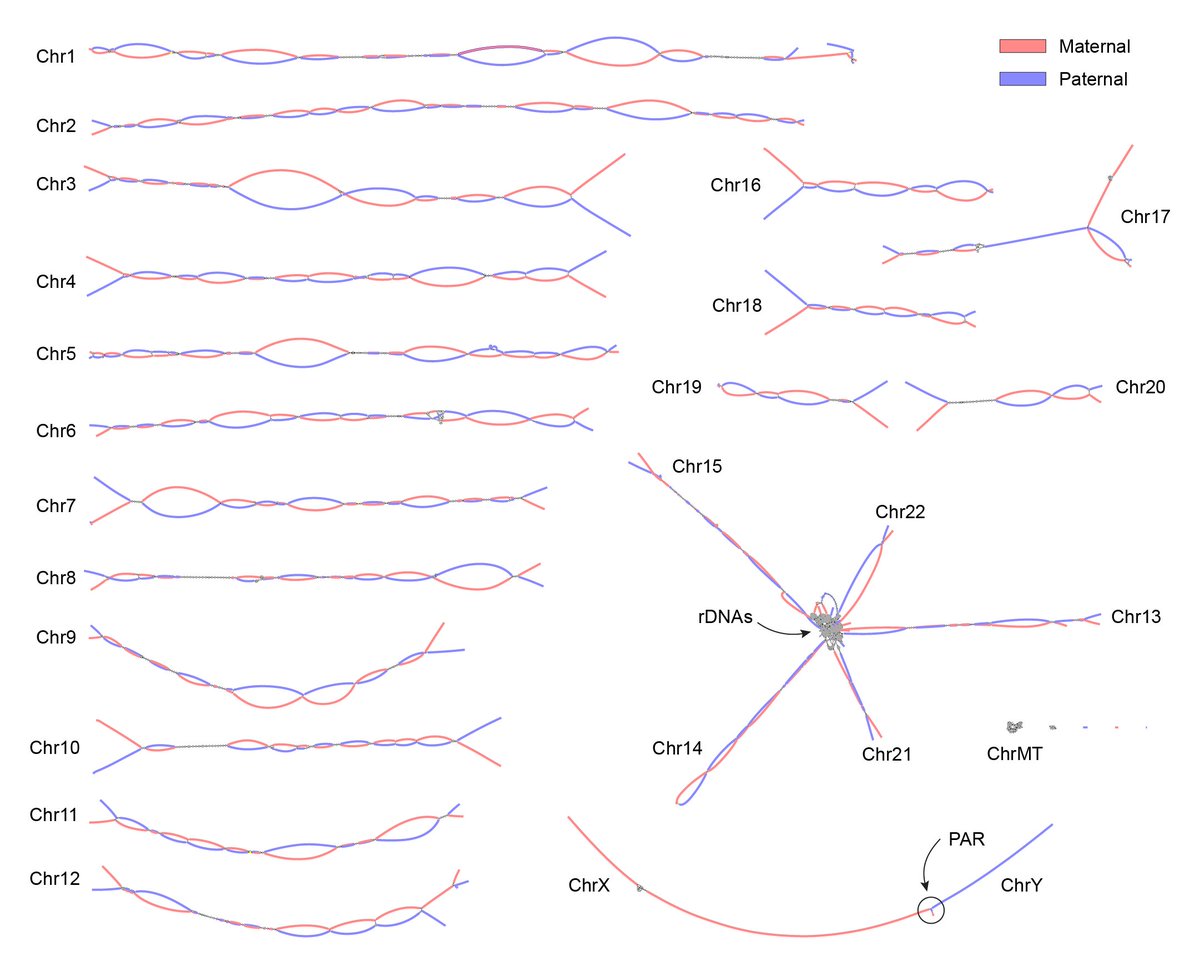
Our next big thing! 🎉 "Verkko: telomere-to-telomere assembly of diploid chromosomes" from Mikko Rautiainen Sergey Nurk 🇺🇦 Sergey Koren generates beautiful graphs and automatically assembles 20/46 human chromosomes from HG002 #T2T without gaps biorxiv.org/content/10.110…

Happy to share our latest work from Winston Timp lab, modbamtools: Analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering 👉manual and tutorial: rrazaghi.github.io/modbamtools/ 👉preprint: biorxiv.org/content/10.110… some of the tools to highlight: 1/

Super stoked to be joining MoGen University of Toronto as an Assistant Professor in Oct '22! I'm founding "The Laboratory for RNA-Based Lifeforms" to explore the diversity of Earth's RNA entities 🧬🔎🦠🌎! Looking to hire🥰! (Please RT) #RNA #genetics #virology #bioinformatics 1/3


The Donnelly Centre for Cellular and Biomolecular Research at the Univ of Toronto is inviting applications for three tenure track assistant professors. These are broad searches in the area of functional genomics (2x) and computational biology 1/2 thedonnellycentre.utoronto.ca/current-opport…


Congratulations to Dr. Heather Gibling (aka Dr! Heather Gibling) for an amazing PhD defence today! Thank you Titus Brown, Moses Lab, Jüri Reimand and Pugh Lab for being on the examination committee. Great to work with you and Awadalla Lab on the most challenging parts of the genome

