Daniel Howrigan
@danhowrigan
Human genetics & reproducible science
ID:1082133710
12-01-2013 07:14:55
855 Tweets
404 Followers
445 Following

GUIDE deconstructs genetic architectures using association studies biorxiv.org/cgi/content/sh… #biorxiv_genetic



🚨🔊Interested in structural variants? Genome sequencing? Pediatric cancers? Germline variants+cancer risk? Rare variants+rare diseases?
If so (or even if not!), I'm delighted to share the first paper from my postdoc Eli Van Allen Dana-Farber:
biorxiv.org/content/10.110…
1/12


Would you believe me if I told you that a single variant in a non-coding RNA explains ~0.5% of all undiagnosed individuals with neurodevelopmental disorders (NDD) in Genomics England ???
I didn’t initially either, but here is the story of RNU4-2 🧵1/9



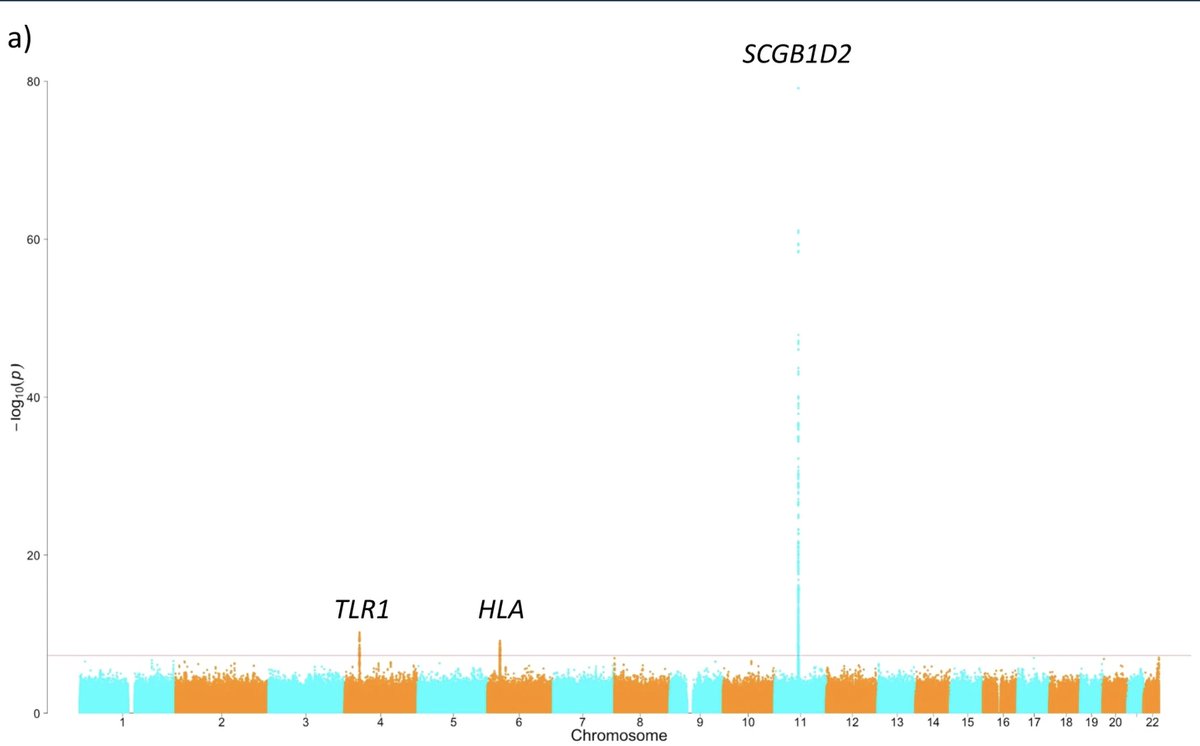
Infertility affects 1 in 6 couples across the globe, but its cause remains unknown in over 30% of cases 😔 In our latest work with Cecilia Lindgren, we discover rare and common genetic variants associated with infertility in 🇬🇧🇩🇰🇪🇪🇫🇮🇮🇸 medrxiv.org/content/10.110… Thread ⬇️

I remember reading this in 2022 as a preprint. Glad to see this work by Satu Strausz, Michal Tal, PhD, Hanna M Ollila and team in published form. This is a nice example of a low-hanging fruit that didn't get picked early because no one studied the phenotype. Performing a GWAS of Lyme









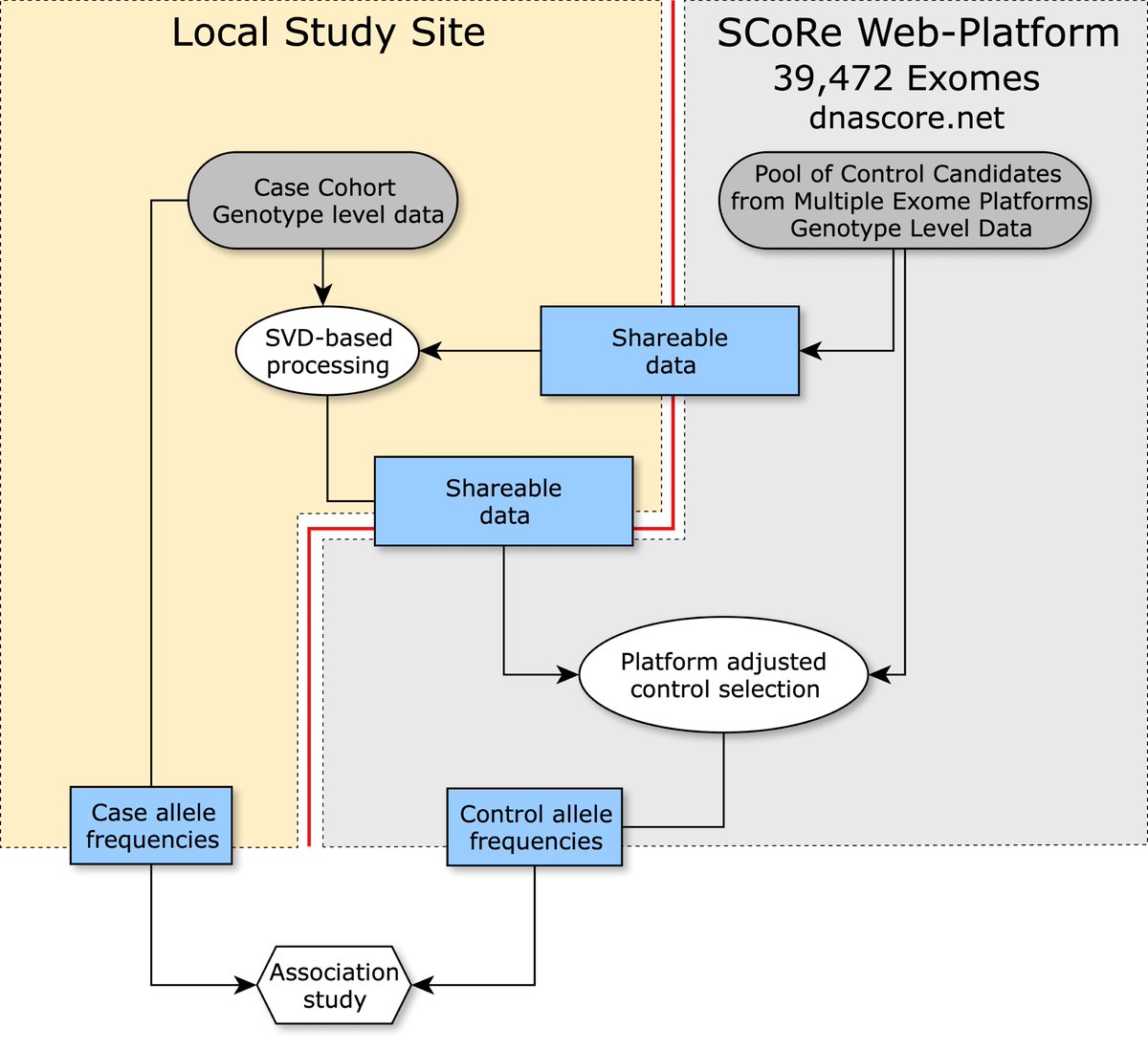
Our team - Alex, Maxim Artyomov and Mark Daly has developed the algorithm for genetic association studies without individual-level data sharing and a resource with 39,472 exomes to be used as a pool of controls. It is now available at Nature Genetics! nature.com/articles/s4158…

New paper by Jiacheng Miao! We often want to study traits that are difficult/expensive to measure. It's becoming popular to use machine learning to predict these phenotypes in large cohorts, then perform GWAS on imputed outcomes. It may be a bad idea!
medrxiv.org/content/10.110…

Our paper on the development of a human genetics-guided score to prioritize drug target genes is online now in Nature Genetics Nature Genetics
Paper: nature.com/articles/s4158…
Share link: rdcu.be/du2ZR
Press release: mountsinai.org/about/newsroom…
1/n