Javier de la Fuente
@Dr_delaFuente
Research Associate @UTAustin | Aging epidemiologist | Genomics | R enthusiast
19-06-2018 09:37:36
309 Tweets
323 Followers
587 Following










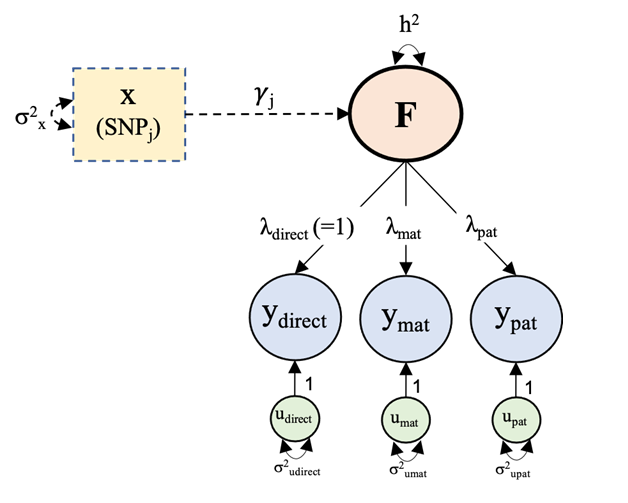
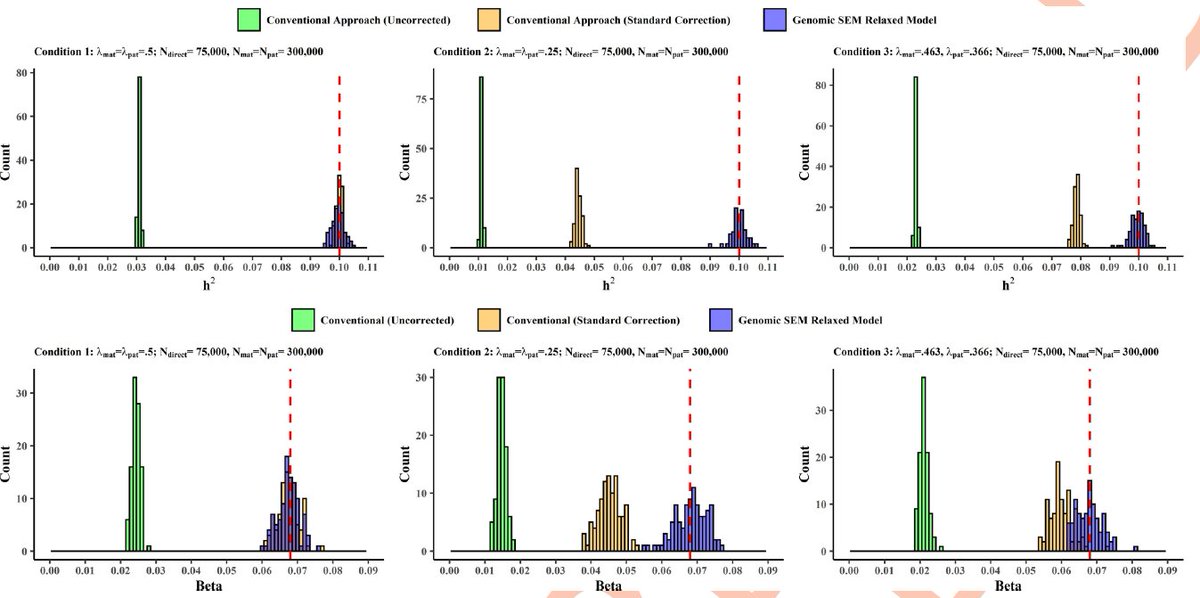
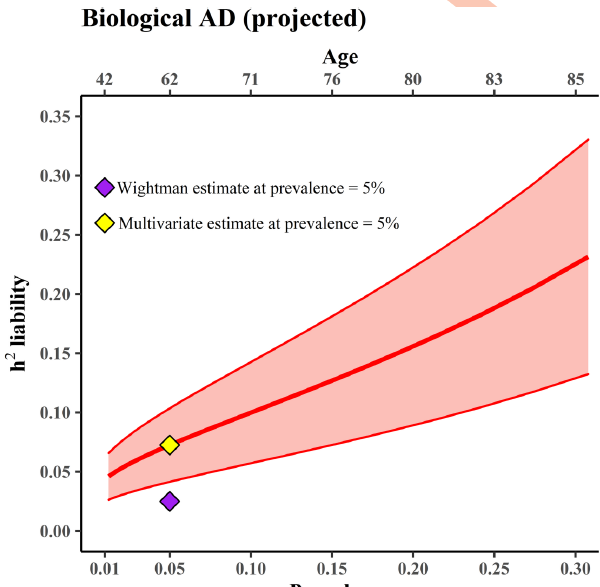
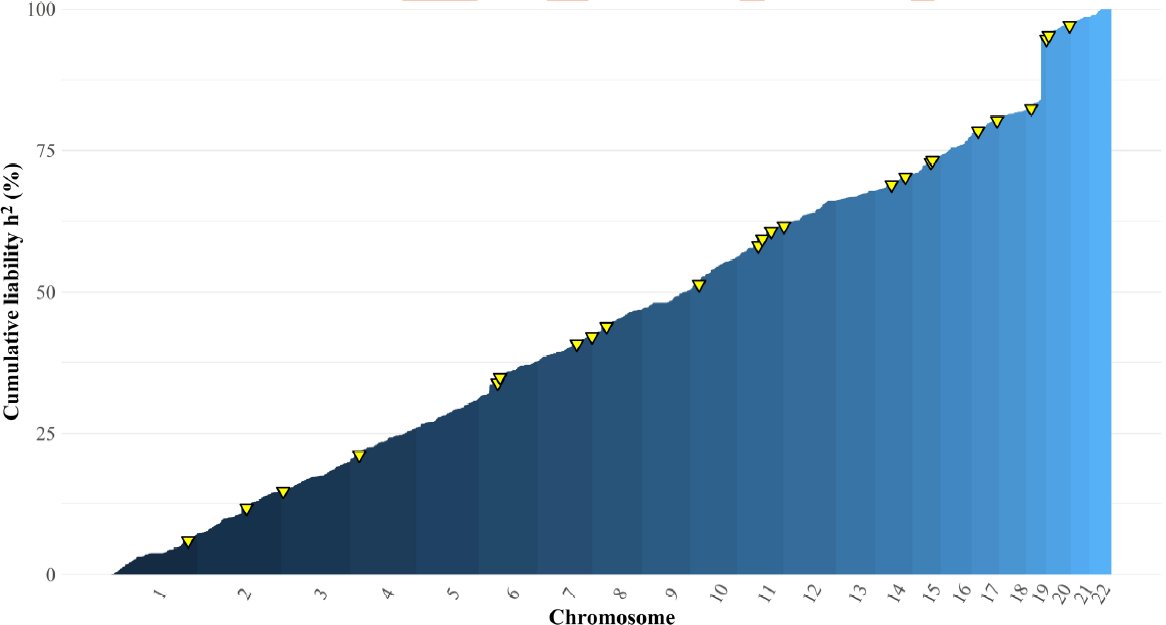
Huge thanks to the amazing team that made this study possible @Andrew_Grotz riccardomarioni @michelnivard Elliot Tucker-Drob 10/10